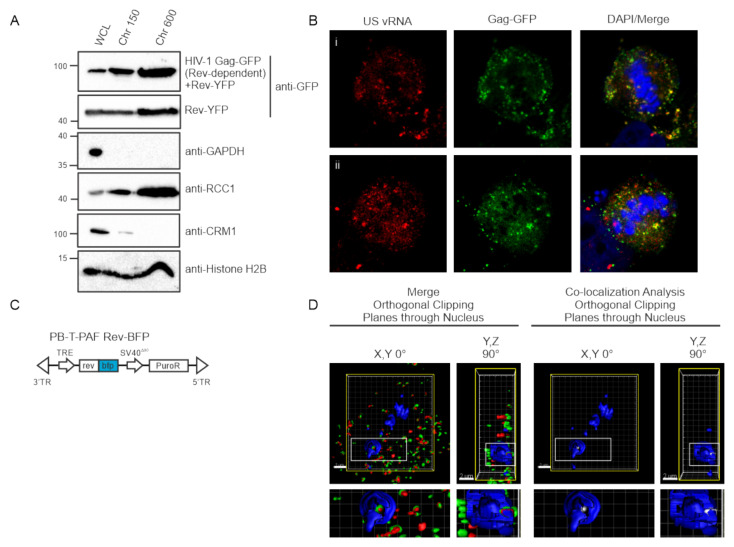
Figure 7.
HIV-1 Gag and Rev are both present in chromatin-associated fractions and are co-localized in three dimensions. (A) 293T cells transfected with HIV-1 Gag-GFP with or without Rev-YFP expression plasmids were separated into chromatin-associated protein fractions representing euchromatin (Chr 150) and heterochromatin (Chr 600) and analyzed by immunoblotting using anti-GFP antibodies. Fraction purity was assessed by immunoblotting with antibodies to detect GAPDH (present only in whole cell lysate, WCL), CRM1 (WCL and chromatin), RCC1 (WCL and chromatin) and Histone H2B (WCL and chromatin). (B) Localization of HIV-1 Gag-GFP (green) and USvRNA (red, detected by smFISH) in two dividing cells (i and ii) showing persistence of Gag-USvRNA co-localization during metaphase but no binding to the chromosome, which was stained blue using DAPI. (C) Schematic diagram of dox-inducible Rev-BFP used to create HeLa HIV.Gag-GFP rtTA Rev-BFP cells. (D) 3D-surface renderings of induced HeLa HIV.Gag-GFP rtTA Rev-BFP cells depicting three-way co-localization in merged images of Gag-GFP (green) and USvRNA (red, detected by smFISH) within the Rev-BFP (blue) mask using orthogonal clipping planes through the nucleus of 0° and 90° rotation. The large white square is the frame that denotes the cell as three-dimensional, the yellow square indicates that the cell was subjected to the orthogonal clipping plane, and the small white rectangle represents the region of interest that was enlarged in the panel below. A co-localization algorithm was used to demonstrate Gag and USvRNA co-localization (white foci) within the confines of the Rev (blue) signal. The small rectangle was enlarged below to provide additional detail.