Figure 5. The ALG14 Hairpin Is Sufficient to Target LDs and It Relies on Similar LD Targeting Features as LiveDrop.
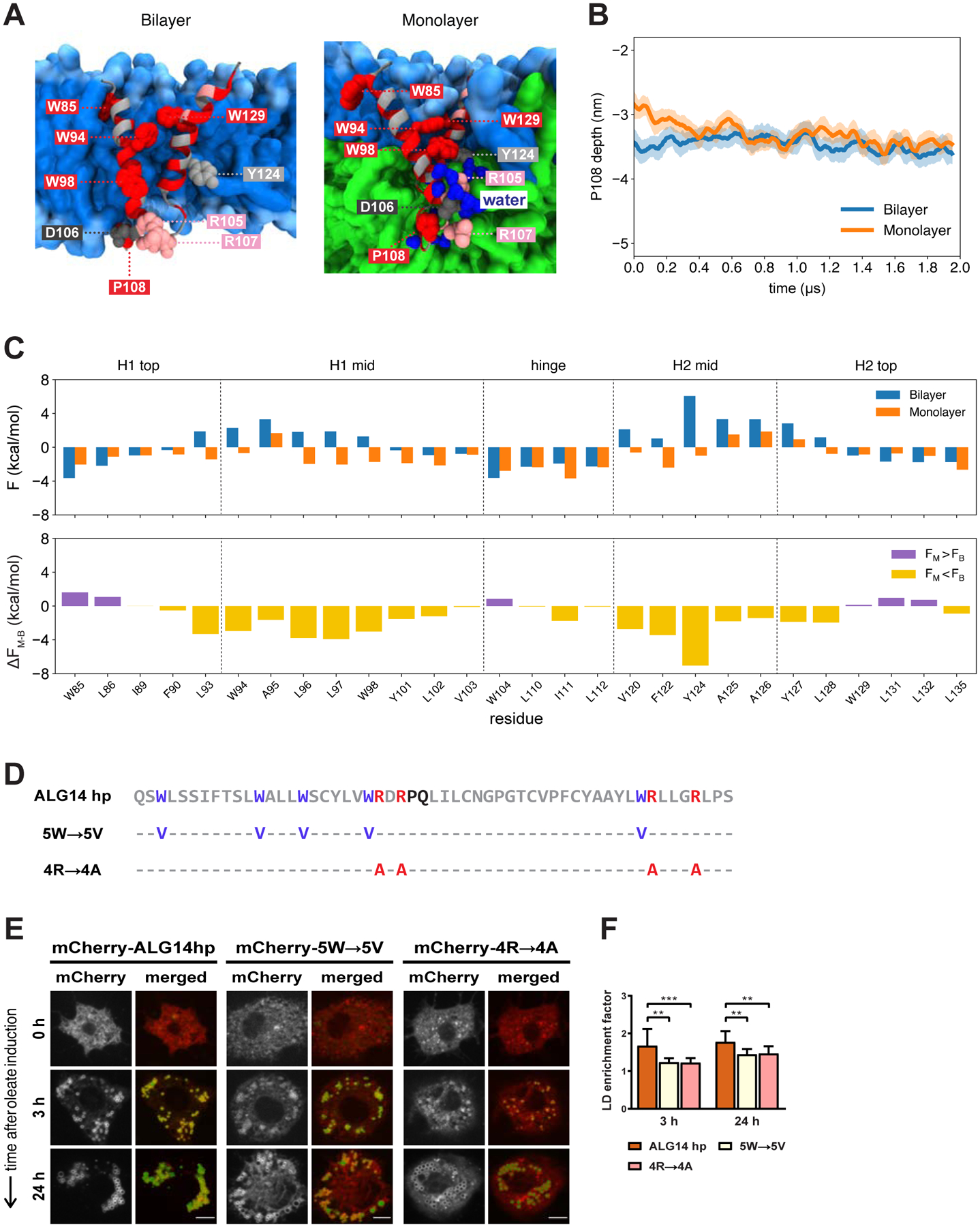
(A) Representative structures of the ALG14 hairpin in a bilayer (left) and monolayer (right) highlighting key residues: R105 and R107 (pink); D106 (dark gray); W85, W94, W98, W129, and P108 (red); and Y124 (light gray). The phospholipid headgroups and tails are shown in light and medium blue, respectively, the neutral lipid phase in green, and water molecules in dark blue.
(B) P108 depth with respect to the upper phosphate plane of the bilayer (blue) and monolayer (orange). The solid lines show the running averages and the filled areas show the standard deviations from four simulations of both the bilayer and monolayer, respectively.
(C) Same as Figure 3C for the ALG14 hairpin.
(D) Amino acid sequence of the ALG14 hairpin (ALG14 hp, gray) and its sequence variants in which the tryptophan (5W→5V, purple) and positively charged (4R→4A, red) residues are individually mutated to valines and alanines, respectively. The predicted hinge of the ALG14 hairpin sequence is shown in black. Amino acid positions indicated with a hyphen (−) remain the same as in the original sequence.
(E) Both of the ALG14 hairpin sequence variants with mutated tryptophans (5W→5V) or positively charged arginines (4R→4A) are compromised in LD accumulation. S2 cells transfected with mCherry-tagged versions of the ALG14 hairpin and its variants (red) were incubated with oleate throughout the indicated time points and imaged by confocal microscopy. LDs were stained with BODIPY (green). Scale bar, 5 μm.
(F) Mean values + SD (n > 13) of the protein signal on LDs after 3 and 24 h of oleate treatment. **, p < 0.01; ***, p < 0.001.
