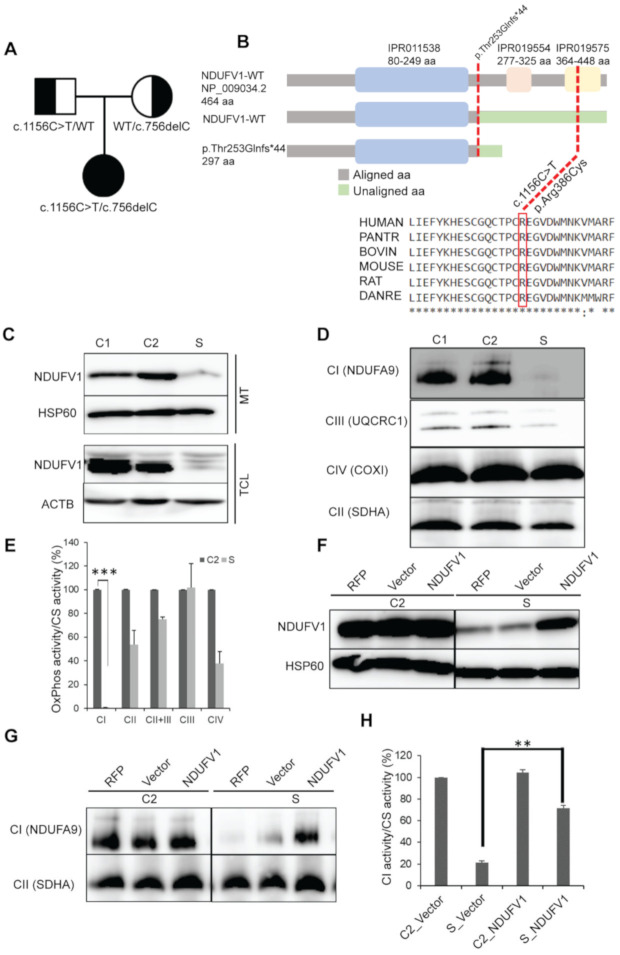
Figure 3.
NDUFV1 mutations detected in a subject. (A) Segregation of the family showing the inheritance pattern of the variants. (B) A graphical representation of NDUFV1 with domains illustrating the position of mutations (red dotted lines) (not to scale). Mutalyzer 2.0.32 (https://mutalyzer.nl/name-checker) for truncated protein prediction, InterPro database (https://www.ebi.ac.uk/interpro/protein) for protein domain prediction, and ClustalW (http://www.ebi.ac.uk/clustalw2) for amino acid (aa) alignment were used. Asterisks (*) indicates conserved aa; IPR011538, NADH: ubiquinone oxidoreductase 51kDa subunit, FMN-binding domain; IPR019554, Soluble ligand binding domain; IPR019575, NADH: ubiquinone oxidoreductase 51kDa subunit, iron-sulfur binding domain. (C) SDS-PAGE immunoblotting of mitochondrial (MT) and total cell lysate (TCL) showing loss of NDUFV1 protein in the subject (S) compared with controls (C1, fHDF; C2, NHDF). HSP60 and ACTB were used as loading controls. (D) BN-PAGE immunoblotting showing destabilization of CI assembly in S. Complex II was used as loading control. (E) OxPhos enzymology revealed CI enzyme deficiency in S compared to C2. Values are expressed as the mean ± SEM. Three independent experiments, two tailed student’s t test done and *** p <0.001 considered statistically significant. (F–H) Lentivirus-mediated complementation assays showing: F, restoration of the NDUFV1 protein in S; G, stabilization of CI assembly; H, restoration of CI enzyme activity. Values are expressed as the mean ± SEM. Three independent experiments, ** p <0.01 considered statistically significant RFP, mtTurbo-RFP; Vector, pCS-CA-MCS.