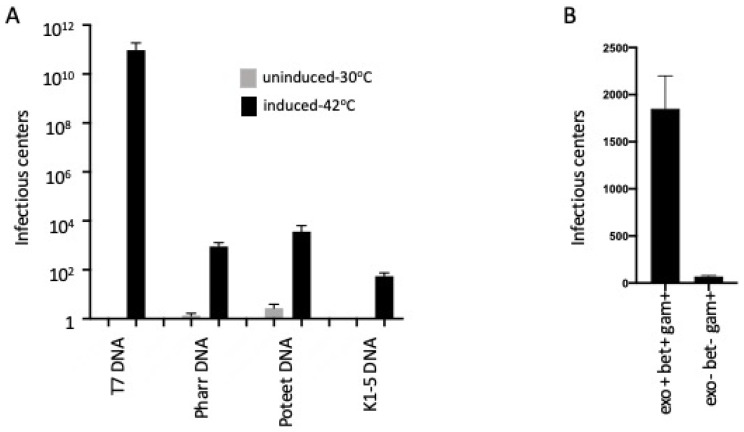
Figure 1.
λ Red function enhances transfection by T7 and T7-related phage genomic DNA. (A) LT976 strain contains a λ defective prophage that expresses Red recombination functions only when induced at 42 °C (see Table 1). The light-colored bars indicate cells grown at 32 °C without Red expression; the dark-colored bars indicate cells induced at 42 °C to express Red recombination functions. Standard error of the mean value is shown and indicates p < 0.005 in Student t-test, comparing induced samples to uninduced controls. Cells, either induced at 42 °C or maintained at 32 °C, were electroporated with ~50 ng of respective phage DNAs (see X-axis, ~1 phage genome per cell). The phage DNA was introduced into LT976 by electroporation as described in Materials and methods. The Y-axis indicates total number of infectious centers per electroporation reaction. The phages Pharr and Poteet were scored on their cognate host Klebsiella pneumoniae 1760b, and the phage K1-5 was scored on its cognate host E. coli K1 (see Table 1). Note that without induction of Red (32 °C no transfectants were found for phage K1-5. (B) In a separate experiment, the RecA defective strain DH5α carries the plasmid pSIM18 or pSIM28. These plasmids contain a defective prophage with genes to express Exo, Beta and Gam or just Gam, respectively. Expression is induced by a shift to 42°. Transfection of the T7 genome is as described in Methods. Bars show level of phage yields following transfection. Negligible infective centers are found in the absence of Exo and Beta.