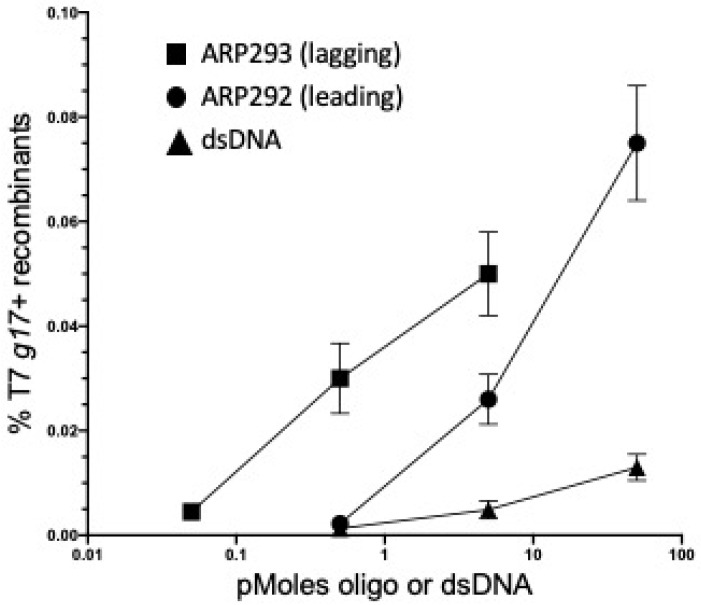
Figure 3.
Recombineering can correct a point mutation following transfection of the T7 genome. The T7 g17am267 mutant genomic DNA (50 ng) was co-electroporated with the substrate DNAs (pmole amounts on X-axis) into the amber suppressor strain ARP3771 expressing the λ Red functions. Single-strand oligonucleotide substrates ARP292 (circles) and ARP293 (rectangles) of 71 nucleotides in length carry the wild type coding information. Their sequences correspond, respectively, to the leading and lagging strands used in T7 DNA replication. The dsDNA substrate (triangles) is a PCR product corresponding in length and sequence to the single-stranded oligos used here. T7 g17+ wild type recombinant frequencies (Y-axis) were calculated by the following equation: (phage titer on E. coli B)/(phage titer on E. coli B supD). Values represent an average of at least three biological replicates with standard errors of the mean indicated. A control T7 genome transfection without added DNA substrate generated no wild type T7 revertants among an average of 4.4 × 104 total plaques recovered on E. coli B supD in eight experiments, indicating a reversion frequency < 2.3 × 10−5.