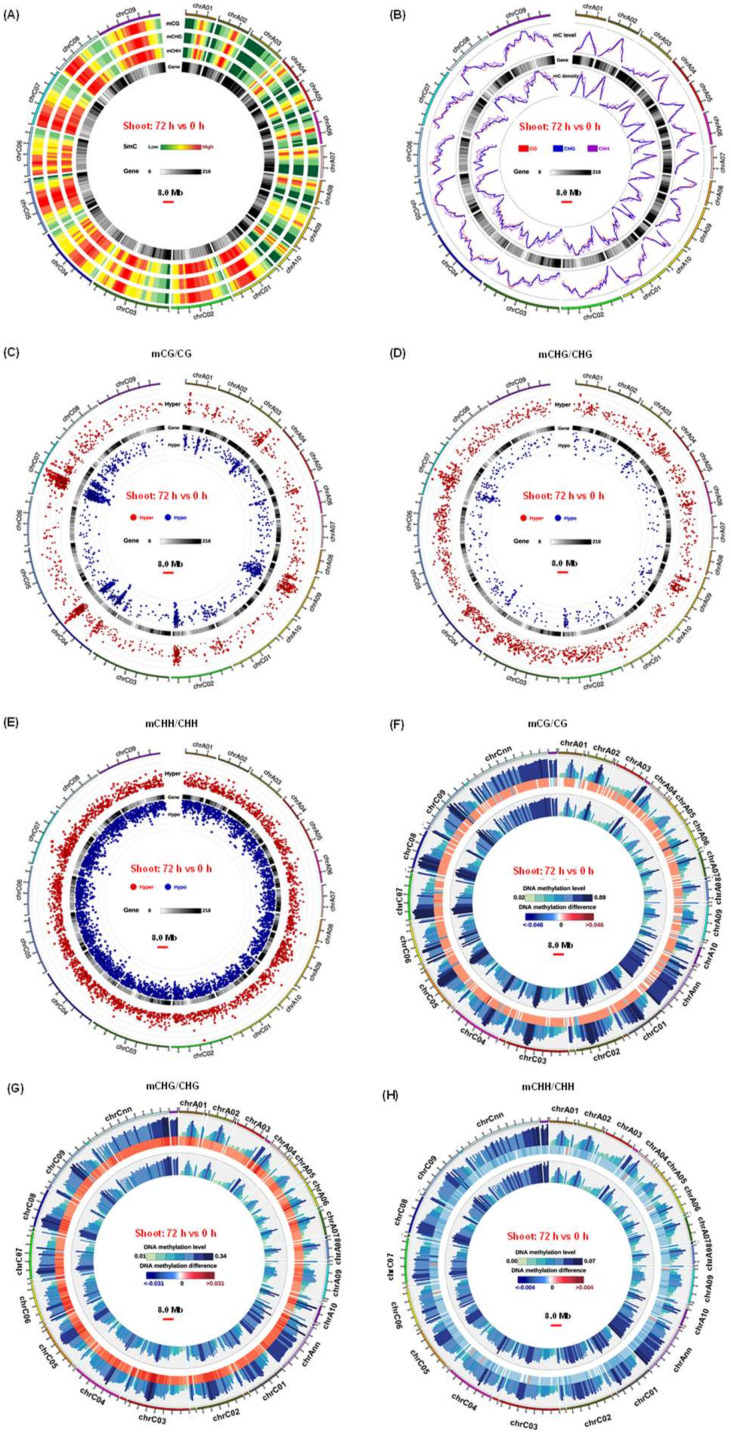
Figure 3.
Overview of the genome-wide differential DNA methylation fingerprints in the rapeseed plant shoots between N sufficiency and N limitation. (A) Circos plots showing the density of differentially methylated regions across the rapeseed genome. In the Circos figure, the terms were as follows outside-to-inside: (i) chromosome, (ii) 5mC density in the CG, (iii) CHG, and (iv) CHH contexts, and (v) gene density of each chromosome in alloteraploid rapeseed. (B) Density and level plot of 5mC in CG, CHG, and CHH contexts in the gene bodies on each chromosome. In the Circos figure, the terms were as follows outside-to-inside: (i) chromosome, (ii) 5mC levels in the CG, CHG, and CHH contexts, (iii) gene density, and (iv) 5 mC densities in the CG, CHG, and CHH contexts in the gene bodies. (C–E) Density of hypermethylated/hypomethylated regions in the (C) CG, (D) CHG, and (E) CHH contexts across the rapeseed genome. In the Circos figure, the terms were as follows outside-to-inside: (i) chromosome, (ii) densities of hypermethylated regions in the CG, CHG, and CHH contexts, (iii) gene density, and (iv) densities of hypomethylated regions in the CG, CHG, and CHH contexts. (F–H) Differential methylation levels in the (F) CG, (G) CHG, and (H) CHH contexts across the rapeseed genome. In the Circos figure, the terms were as follows outside-to-inside: (i) chromosome, (ii) hypermethylation levels in the CG, CHG, and CHH contexts, (iii) DNA methylation differences, and (iv) hypermethylation levels in the CG, CHG, and CHH contexts. The times of 0 h and 72 h refer to the times after treatment of N deficiency.