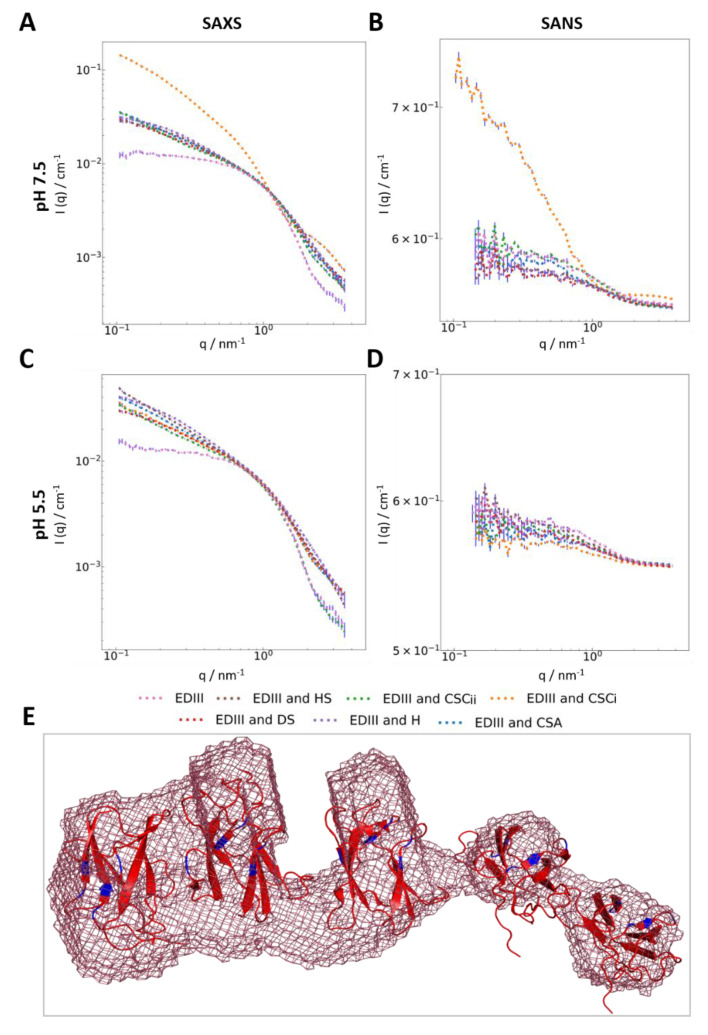
Figure 4.
Scattering data for EDIII with GAGs at pH 7.5 and 5.5, and modelling of EDIII with CSCi. (A) SAXS data of EDIII with GAGs at pH 7.5, (B) SANS data of EDIII with GAGs at pH 7.5, (C) SAXS data of EDIII with GAGs at pH 5.5, (D) SANS data of EDIII with GAGs at pH 5.5, after subtraction of background, empty cell, and incoherent scattering. Error bars shown in blue. (E) Ab initio modelling of low-resolution SANS data of EDIII with CSCi at pH 7.5 using DAMMIF, χ2 = 0.9527 [32] represented as a light red mesh with X-ray structures of EDIII monomers (PDB code 6FLA) placed in areas of larger electron density. EDIII residues previously identified as potential GAG binding sites are shown in blue [6].