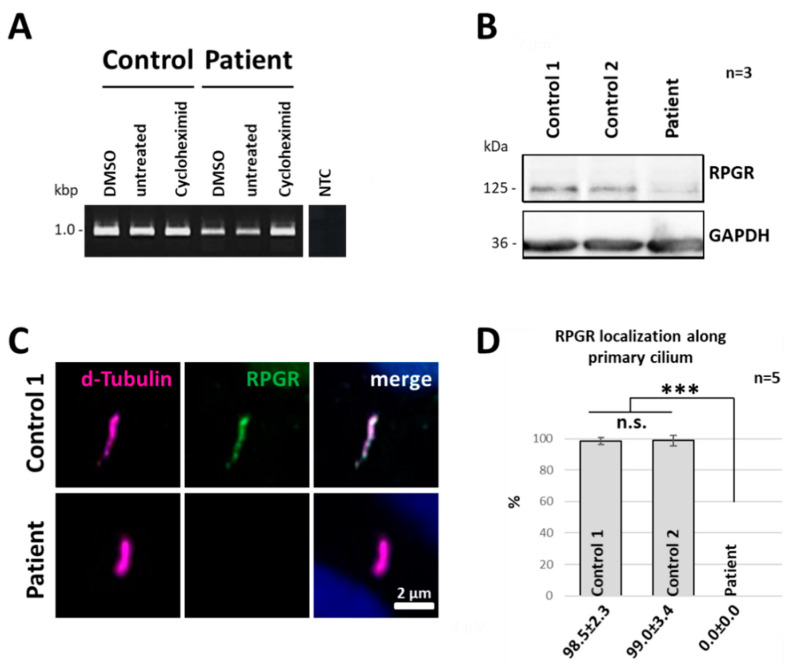
Figure 4.
Patient-derived dermal fibroblasts lack RPGR at primary cilia. (A) Influence of NMD on RPGR transcript levels in patient-derived and control fibroblasts. Cells were treated with 100 µg/mL cycloheximide (CHX) for 4 h. DMSO-treated and untreated cells were used as controls. RT-PCR analyses included exon 9 to 11 of the RPGR transcript and were performed using equal amounts of RNA in each sample. NTC: non-template control. (B) Western blot analyses showed a strongly reduced level of RPGR protein expression (125 kDa) in the patient-derived cell line. Residual signals in the patient might correspond to basal spontaneous read-through. In contrast, RPGR was detected in two unaffected control cell lines. GAPDH (36 kDa) was used as a loading control. Molecular weight is indicated in kDa. (C) Immunocytochemistry (ICC) of RPGR at primary cilia. Control cells showed a dot-like expression pattern of RPGR (green) along the axoneme of the cilium (magenta). In the patient-derived fibroblasts, RPGR expression was not detected. D-tubulin was used as a marker for the primary cilium. (D) Quantification of cilia showing RPGR along the ciliary axoneme. A highly significant reduction of RPGR signals at primary cilia was detected in the patient-derived cell line. More than 90% of the control cells showed RPGR at the primary cilium, whereas RPGR was not detected along the cilium in the patient cell line (among 250 primary cilia, none showed RPGR staining). Experiments were independently repeated 5 times (n = 5). Error bars represent standard deviation. Asterisks: significant differences; n.s: non-significant, ***: p ≤ 0.001.