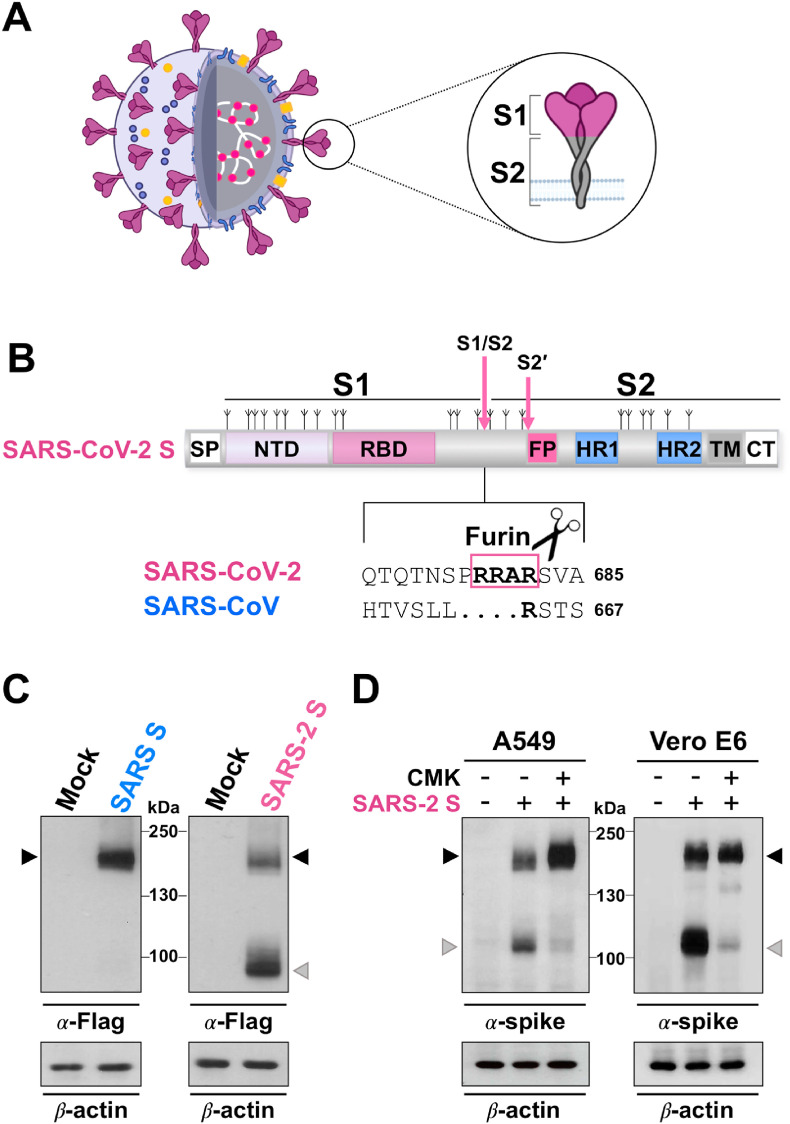
Fig. 1.
Furin-mediated S1/S2 cleavage during SARS-CoV-2 spike biogenesis. (A) Schematic representation of the SARS-CoV-2 virus and the spike glycoprotein. The lipid bilayer comprising the spike protein (S, violet), the membrane protein (M, blue) and the envelope protein (E, orange), and the viral RNA (white) associated with the nucleocapsid protein (N, pink) are shown. The spike protein S1 and S2 subunits are indicated in the zoom. (B) Schematic representation of the SARS-CoV-2 S glycoprotein. The positions of N-linked glycosylation sequons are shown as branches. S1, receptor-binding subunit; S2, membrane fusion subunit. The S1/S2 and S2’ protease cleavage sites are indicated by arrows. Protein domains are illustrated: SP, signal peptide; NTD, N-terminal domain; RBD, receptor binding domain; FP, fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Sequence comparison of the S1/S2 cleavage site of SARS-CoV-2 and SARS-CoV spike proteins, and the putative furin cleavage site (RRAR residues in the box) in SARS-CoV-2 are shown. GenBank accession numbers are QHD43416.1 for SARS-CoV-2 spike and AFR58740.1 for SARS-CoV spike [104]. (C) Human A549 alveolar type II-like epithelial cells (ATCC) transfected with Flag-tagged SARS-CoV-2 spike (SARS-2 S, C-terminal Flag VG40590-CF, Sino Biological) or Flag-tagged SARS-CoV spike (SARS S, C-terminal Flag VG40150-CF, Sino Biological) constructs, or empty vector (Mock) were analyzed by immunoblot (IB) using anti-Flag antibodies (Cell Signaling). (D) IB for SARS-CoV-2 spike protein using anti-SARS-CoV-2 spike antibody (α-spike, Sino Biological) in whole cell extracts from A549 and Vero E6 cells (ATCC) transfected with the SARS-CoV-2 spike construct (SARS-2 S, +) or empty vector (−), and treated with the furin inhibitor decanoyl-RVKR-CMK (CMK, Sigma-Aldrich) or vehicle (−). In panels C and D, black arrowheads indicate bands corresponding to uncleaved S protein, whereas gray arrowheads indicate the cleaved S fragment.