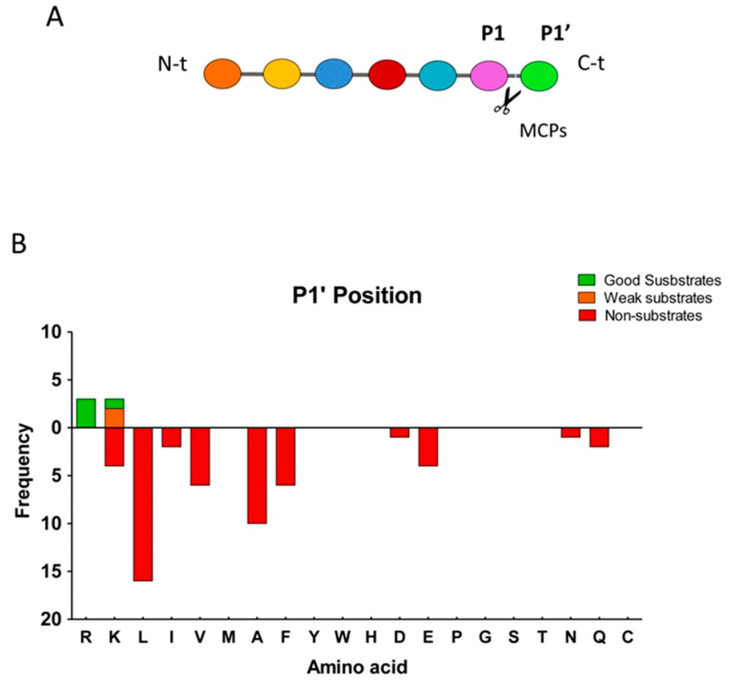
Figure 4.
Analysis of the substrate preferences of CPZ from the peptidomics experiment using the HEK293T-derived peptide library. (A) Schematic representation of relevant residues involved in a typical MCPs cleavage, according to the model proposed by Schechter and Berger [32]; (B) Substrate preference of CPZ at the C-terminal (P1′) position. The frequency for each amino acid found in P1′ is indicated for good substrates, weak substrates, and non-substrates.