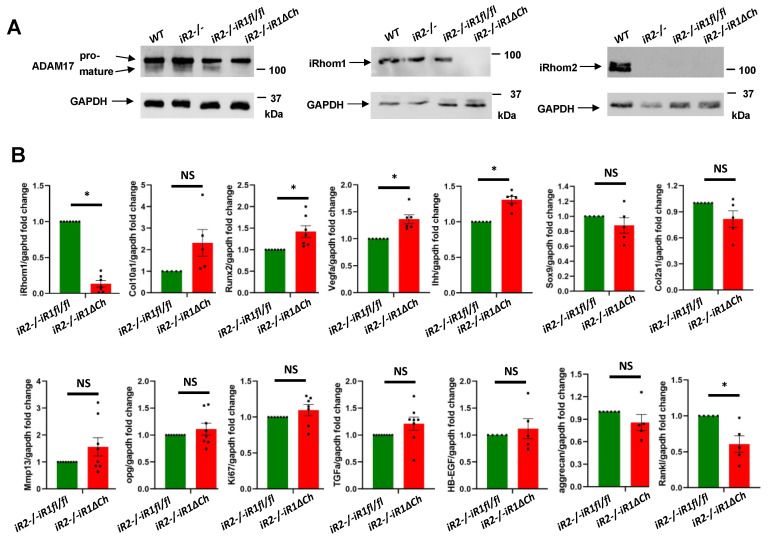
Figure 5.
Western blot analysis of ADAM17, iRhom1 and iRhom2 expression and qPCR analysis for chondrocyte and bone markers. (A) Western blots of samples prepared from primary chondrocytes from WT, iR2−/−, iR2−/−iR1fl/fl or iR2−/−iR1∆Ch animals were probed for ADAM17 (left panels, see Supplementary Figure S7 for quantification), iRhom1 (middle panels) or iRhom2 (right panels). The Western blots confirmed the presence of pro-ADAM17 in all samples, whereas mature ADAM17 could only be detected in WT, iR2−/− and iR2−/−iR1fl/fl samples, but not in iR2−/−iR1∆Ch chondrocytes (left panels). A Western blot for iRhom1 demonstrated that it was present in primary chondrocytes from WT, iR2−/− and iR2−/−iR1fl/fl mice but had been efficiently deleted in iR2−/−iR1∆Ch chondrocytes (middle panels). iRhom2 could only be detected in WT primary chondrocytes, but not in chondrocytes from iR2−/−, iR2−/−iR1fl/fl or iR2−/−iR1∆Ch mice (right panels). In all cases, GAPDH served as a loading control. Each Western blot is representative of at least 3 separate experiments. (B) qPCR analysis showed an almost abolished expression of iRhom1 in iR2−/−iR1∆Ch chondrocytes (top left graph). There was a non-significant trend towards increased expression of Col10a1, increased expression of Runx2, Vegfa and Ihh, no significant difference in the expression of Sox9, Col2a1, Mmp13, OPG, Ki67, TGFα, HB-EGF or aggrecan, and reduced expression of Rankl in iR2−/−iR1∆Ch samples compared to iR2−/−iR1fl/fl controls. Each qPCR analysis was performed on at least 3 samples of each genotype. * indicates statistical significance with a p-value of <0.05 in a student’s t-test, whereas NS indicates no significant difference. The SEM is indicated by error bars.