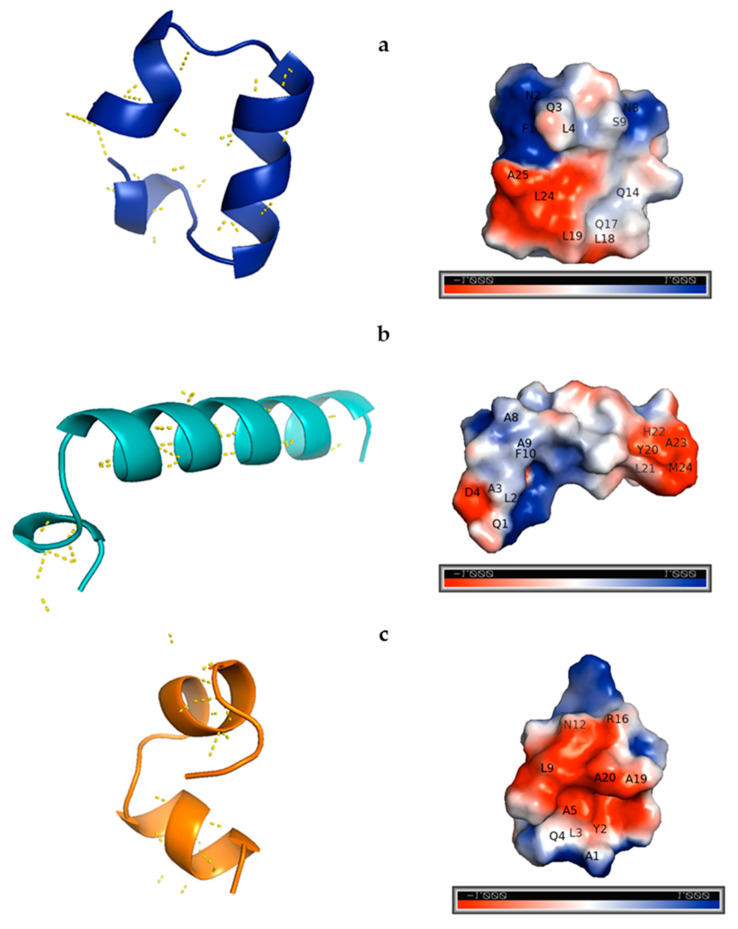
Figure 1.
Left side: predicted 3D structure of the synthesized peptides and polar interactions between amino acids. Right side: electrostatic potential according to the predicted structures of the synthesized peptides. The QUARK online Ab initio protein structure algorithm was used to obtain the models and PyMOL molecular graphics system version 2.3.4 was used to generate the 3D model images. The Adaptive Poisson–Boltzmann Solver (APBS) plugin version 3.7 was used to calculate and predict the electrostatic potential surface. The electrostatic potential scale ranges from −1 to +1. Amino acids contributing with the potential are represented as one letter codes and their number in the sequence. (a) 19ZP1; (b) 19ZP2; (c) 19ZP3.