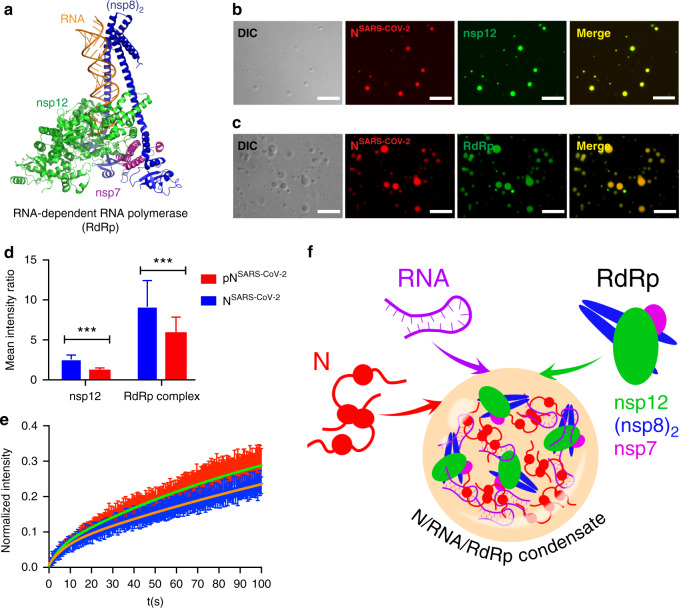
Fig. 5. The RNA-dependent RNA polymerase complex of SARS-CoV-2 concentrates in RNA/nucleocapsid protein-droplets.
a Structure of the RNA-bound RdRp-complex formed by the SARS-CoV-2 non-structural proteins nsp12 (green), nsp7 (magenta) and nsp8 (blue) in 1:1:2 stoichiometry (PDB code: 6YYT)29. b Fluorescence and DIC microscopy show the recruitment of Alexa Fluor 488 labeled nsp12 (green), the catalytic subunit of the RdRp-complex, into NSARS-CoV-2/polyU droplets (NSARS-CoV-2 in red). c Active SARS-CoV-2 RdRp-complex bound to a fluorescein-labeled minimal RNA hairpin concentrates inside of NSARS-CoV-2/polyU droplets. In (b) and (c) droplets of 50 µM NSARS-CoV-2 and 1 µM polyU were prepared in 20 mM NaPi, pH 7.5, and visualized by the addition of a small amount of Alexa Fluor 594 labeled NSARS-CoV-2 protein. Scale bars are 10 µm in (b) and (c). Micrographs are representative of three independent biological replicates. d Recruitment of nsp12 and RdRp complex into polyU-induced droplets of unmodified (blue) and SRPK1-phosphorylated (red) NSARS-CoV-2. Mean values and standard deviation are shown (n = 100 droplets). Two-sided t-test with P value set to < 0.05 for statistical significance, ***<0.001, **<0.002 and *<0.033, ns < 0.12. e FRAP of nsp12 after recruitment into NSARS-CoV-2/polyU (blue) and SRPK1-phosphorylated NSARS-CoV-2/polyU (red) droplets. Error bars represent the standard deviation for averaged 10 and 11 curves for unmodified and phosphorylated NSARS-CoV-2, respectively. (f) Schematic representation of the LLPS-based formation of N/RNA/RdRp-condensates as protein/RNA-dense sites for viral transcription.