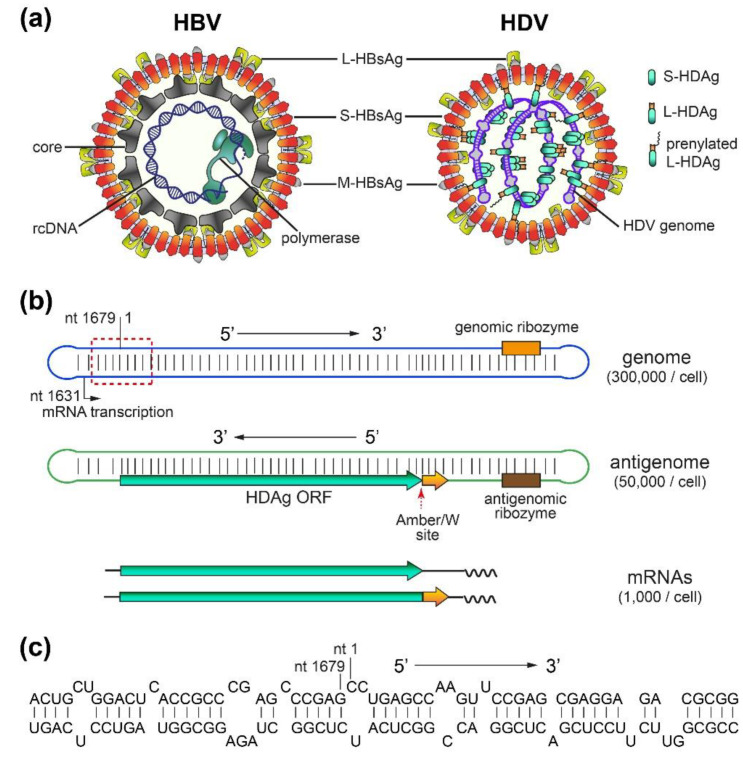
Figure 1.
Hepatitis D virus (HDV) and Hepatitis B virus (HBV) virions, and HDV RNAs. (a) Schematic representation of HDV and HBV virions. Both viruses share the same envelope containing three HBV envelope proteins: large- (L-), medium- (M-), and small- (S-) HBsAg. HDV (right panel) has a ribonucleoprotein (RNP) complex inside. The RNP consists of the HDV genome and two isoforms of hepatitis D antigen (HDAg), L- and S-HDAg. A portion of L-HDAg is prenylated, which is needed for its association with S-HBsAg [35]. On the other hand, HBV (left panel) has a nucleocapsid inside the envelope. The nucleocapsid consists of an HBV core protein shell and relaxed circular HBV DNA (rcDNA), with the latter associated with HBV polymerase. (b) HDV genome, antigenome and mRNAs. The HDV genome is a single-strand, negative-sense, circular RNA. It forms an unbranched rod-like structure due to its high degree of intramolecular base-pairing. The HDV antigenome is complementary to the genome and is predicted to form a similar structure to the genome. Two mRNAs encoding either S-HDAg or L-HDAg are transcribed using the genome as a template. Ribozymes, the ADAR1 editing (Amber/W) site, mRNA transcription starting site, and HDAg open reading frame (ORF) are indicated. Arrows indicate the 5′ to 3′ direction. (c) Structure of a representative region of the genome (red dash line box in (b)) consisting of short stems and bulges.