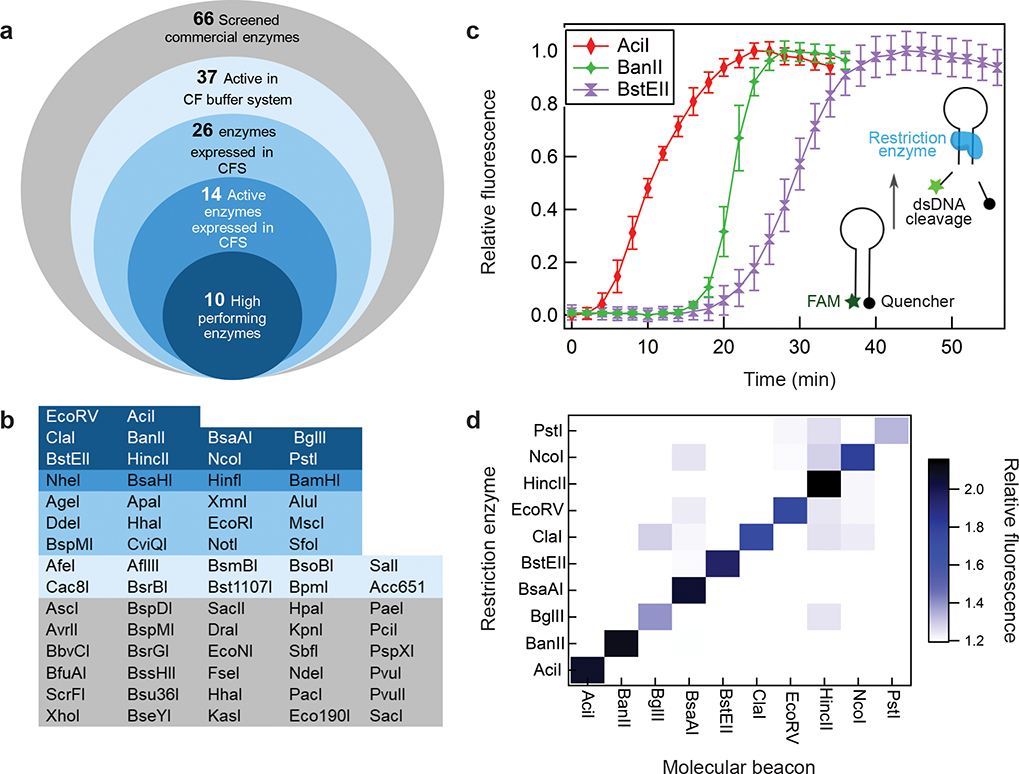
Figure 2. Development of orthogonal, restriction enzyme-based reporters.
a. Candidate restriction enzymes were evaluated through a four-step screening pipeline (i-iv) designed to find enzymes with high rates of expression and processivity in the cell-free expression system (CFS). i) 37 of 66 commercially available enzymes demonstrated activity in the cell-free (CF) buffer system that replicates the pH, buffer and salt composition found in the complete transcription and translation system, ii) 26 of the 37 above restriction enzymes were successfully expressed de novo in the cell-free system (Supplementary Fig. 1), iii) 14 of these 26 cell-free expressed enzymes showed high levels of cleavage activity (Supplementary Fig. 2), iv) 10 of the 14 enzymes demonstrated high rates of enzyme-mediated cleavage from de novo cell-free expression. b. A summary of the performance for screened restriction enzymes with colors matching the categories described in Figure. 2a. c. Representative data of three candidate restriction enzyme-based reporters in molecular beacon cleavage assays. Data presented as percent of maximum fluorescence for each molecular beacon, error bars represent SE (N=3, Supplementary Fig. 3). d. Heat map of specific enzyme activity. All combinations of restriction enzymes and molecular beacons were tested. Values are average of triplicates at 180 min.