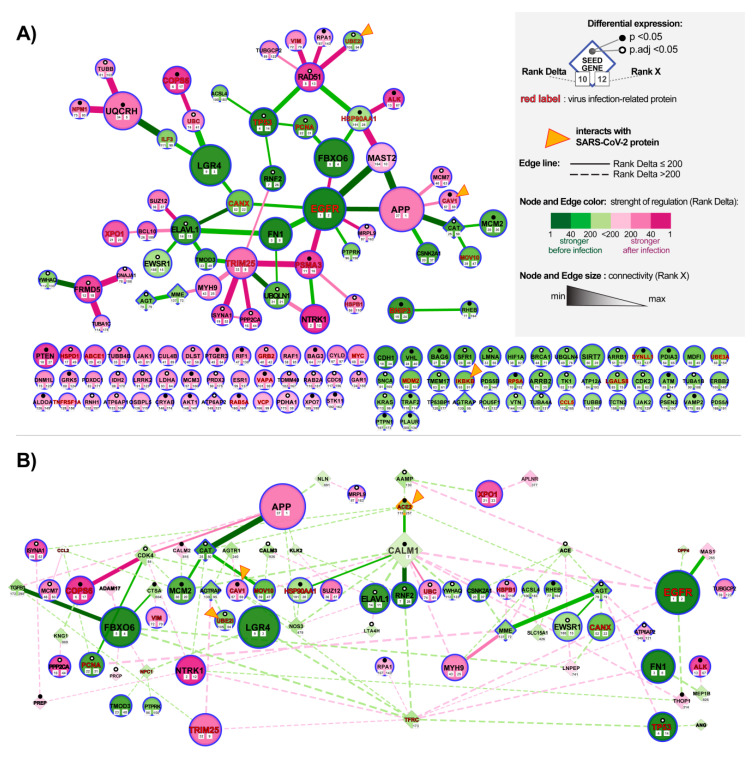
Figure 4.
Alteration of ACE2 (Angiotensin-Converting Enzyme 2) networks in cardiomyocytes infected with SARS-CoV-2. (A) ACE-2 related hub genes with corroborated signaling obtained by analyzing expression data of human-induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) after 72 h of infection with SARS-CoV2. The network was constructed by using the NERI (Network by Relative Importance) algorithm which integrates protein-protein interaction (PPI) BioGrid interactome with gene co-expression network. For clarity, we selected top genes and edges which had Rank Delta and Rank S number between 1 and 200; additionally, we showed nodes with the best NERI scores (low-rank number) which did not have associated edges with best scores (low-rank number). Genes marked with orange triangles showed direct interaction with SARS-Cov-2 [39]. Notice that EGFR (Epidermal Growth Factor Receptor) and APP (Amyloid Beta Precursor Protein) showed the strongest alterations in their co-expression networks. (B) PPI network between top co-expressed hub genes from panel A (circular shapes) and seed genes (diamond shapes) related to the complete ACE2 network identified using data mining. The size of the nodes and weight of the edges is associated with Rank X number, related to biological importance, while color is associated with Rank Delta, related to the difference in co-expression network between control and disease. Notice that ACE2 shows a reduced number of connections, consistent with its initial downregulation in the early stage of infection; in later stages of infection, we can expect an inversion of observed regulation caused by the virus-related ACE2 overexpression [40].