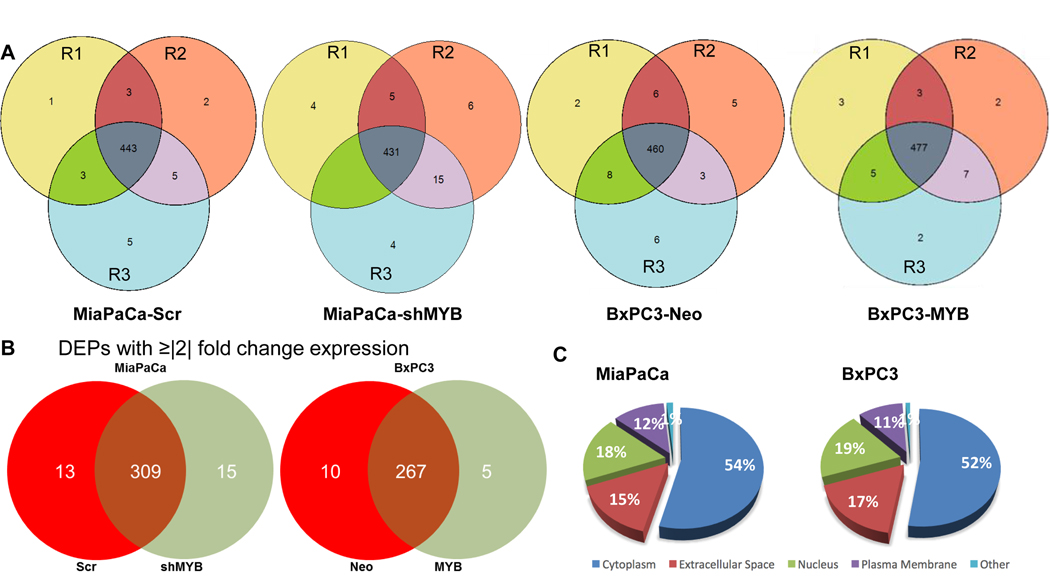
Figure 1: Identification of differentially expressed proteins in the secretome of MiaPaCa (-Scr and -shMYB) and BxPC3 (-Neo and -MYB).
(A) Conditioned media from three populations of MiaPaCa (-Scr and –shMYB), BxPC3 (-Neo and –MYB) differing in passage number, but strictly cultured under similar growth conditions were collected, pooled and processed for LC-MS/MS analysis as mentioned in the “Materials and methods” section. R1, R2, and R3 represent the three technical replicates of three pooled biological replicates. (B) Proteins identified with at least 3 peptide hits with p-value≤ 0.05 and present in at least 2 two out of three technical replicates of MiaPaCa (-Scr and –shMYB) and BxPC3 (-Neo and –MYB) cell lines were selected for the analysis. Data represent the differentially expressed proteins (DEPs) with ≥|2| fold change between paired MYB-modulated pancreatic cancer cell lines. (C) The pie chart represents the classically associated cellular localization of the total DEPs identified in MiaPaCa (-Scr and –shMYB) and BxPC3 (-Neo and –MYB).