FIGURE 4.
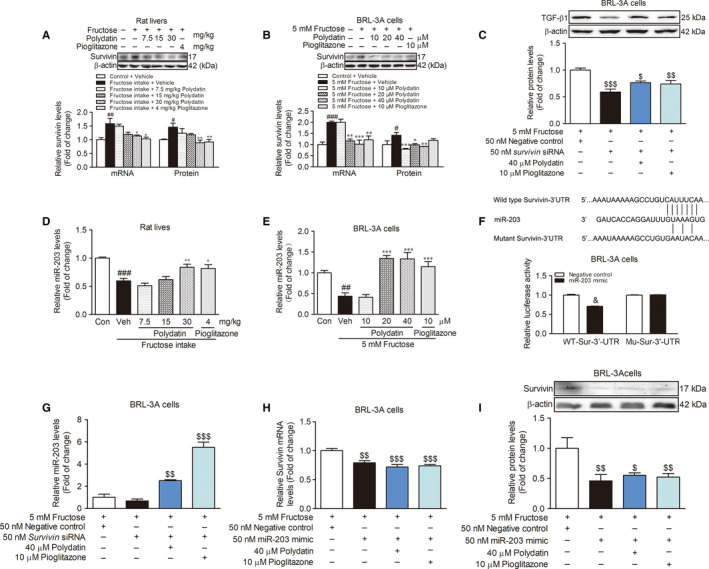
Polydatin augments miR‐203 targeting survivin to inhibit the activation of TGF‐β1/Smad signalling. qRT‐PCR analysis of survivin mRNA levels and Western blot analysis of survivin protein levels in rat livers (A) and in BRL‐3A cells (24 h) (B). (C) TGF‐β1 protein levels (24 h) in transfected with 50 nM survivin siRNA or NC BRL‐3A cells exposed to fructose in the presence or absence of 40 μM polydatin or 10 μM pioglitazone. qRT‐PCR analysis of miR‐203 expression in rat livers (D) and BRL‐3A cells (12 h) (E). (F) The assay of survivin 3′‐UTR luciferase activity. (G) MiR‐203 expression in transfected with 50 nM survivin siRNA or NC BRL‐3A cells treated with fructose in the presence or absence of 40 μM polydatin or 10 μM pioglitazone. U6 was as internal control. Survivin mRNA (H) and protein (I) levels in transfected with 50 nM miR‐203 mimic or NC BRL‐3A cells co‐cultured with fructose in the presence or absence of 40 μM polydatin or 10 μM pioglitazone. β‐actin was as internal control. Each value is shown as mean ± SEM (n = 4‐6). # P < .05, ## P < .01, ### P < .001 compared with the normal control; *P < .05, **P < .01, ***P < .001 compared with the fructose control; $ P < .05, $$ P < .01, $$$ P < .001 compared with the fructose‐negative control; & P < .05 compared with the negative control. UTR, untranslated region
