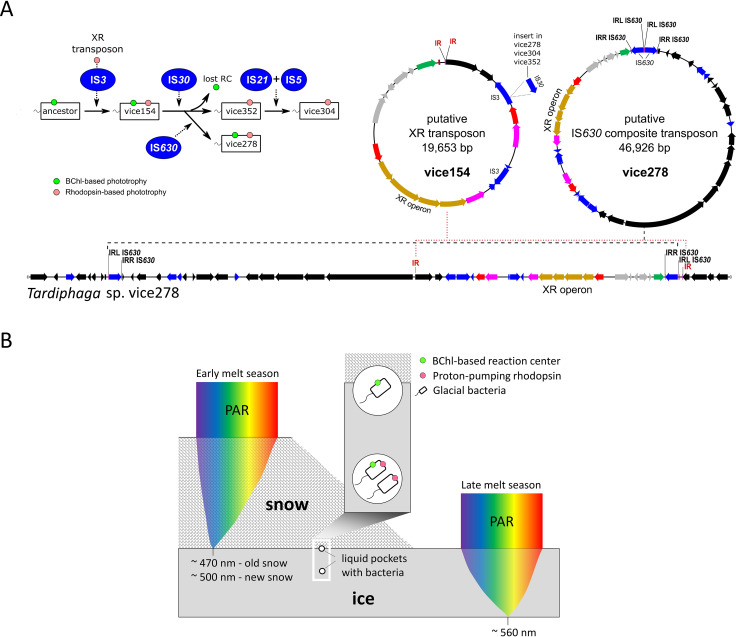
FIG 2.
(A) Hypothetical evolutionary path of the Tardiphaga isolates from their common ancestor based on the IS insertion patterns (left) and two transposons proposed to drive the movement of the xanthorhodopsin (XR) operon (right). The genomic region surrounding the XR operon in Tardiphaga strain vice278 was shown to highlight the distribution of direct or inverted repeats that are required to form the putative transposons. RC, reaction center; IR, inverted repeat; DR, direct repeat, IRL, inverted repeat-left; IRR, inverted repeat-right. (B) A model for light availability in snow and ice and hypothesized niche partitioning of BChl- and rhodopsin-based dual phototrophy versus only BChl- or rhodopsin-based single phototrophy. The snowpack and icepack are depicted to be of such an ideal thickness that the light spectrum with the lowest extinction coefficient reaches the exact bottom of snowpack or icepack. PAR, photosynthetically active radiation. Note that new and old snows have different spectral distribution for PAR.