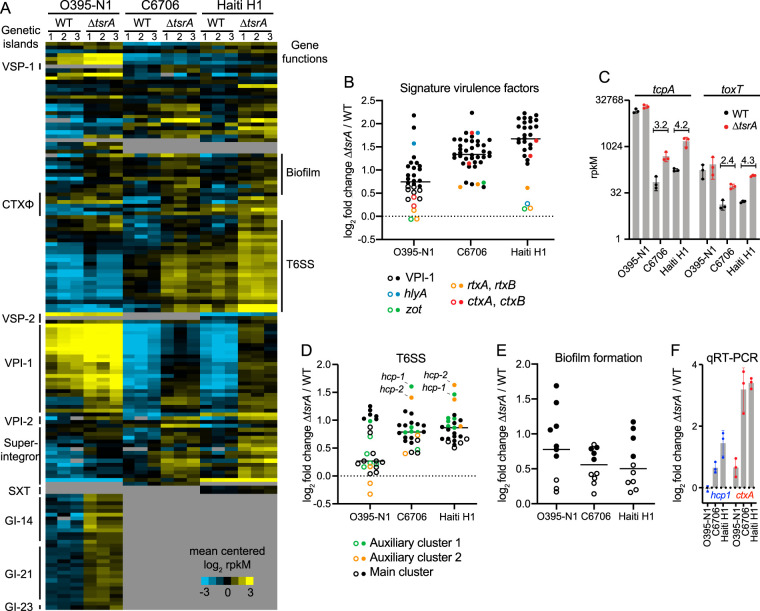
FIG 2.
The majority of genes in the TsrA regulon are encoded on genetic islands that are most highly activated in the Haiti H1 ΔtsrA strain. (A) Heat map of mean-centered log2-transformed expression values of members of the TsrA regulon in each of the three biological replicates of each wild-type and mutant strain. Each row in the heat map corresponds to a gene, and each column in the heat map corresponds to a sample (identified above the heat map); if no ortholog of the gene is found in the strain, the field is shaded gray. Genetic islands are indicated on the left of the heat map, and clusters of biofilm formation and T6SS genes are indicated on the right. (B, D, and E) Fold changes from the wild type in the expression of genes encoding signature virulence factors (B), the T6SS (D), and biofilm formation functions (E) in the ΔtsrA mutant. Filled circles indicate genes considered significantly differentially expressed and part of the TsrA regulon in the corresponding strain (ac, ≥1.3). Open circles indicate genes not regulated by TsrA. Horizontal bars indicate mean values. (C) tcpA and toxT mRNA abundances expressed in reads per kilobase per million (rpkM) (y axis on a log2 scale) in either the WT (black circles) or the ΔtsrA mutant (red circles) of each of the three strains analyzed. Numbers above the C6706 and Haiti H1 data sets indicate the fold rpkM change between the WT and the ΔtsrA mutant. Error bars indicate standard deviations of rpkM measurements for the three independent biological replicates. (F) qRT-PCR measuring the fold change in hcp1 (blue) and ctxA (red) mRNA abundances between the WT and the ΔtsrA mutant in each of the strains analyzed. Error bars represent the standard deviations of measurements obtained from three independent biological replicates.