Abstract
Premise
How genetic variation within a species affects phytochemical composition is a fundamental question in botany. The ratio of two specialized metabolites in Cannabis sativa, tetrahydrocannabinol (THC) and cannabidiol (CBD), can be grouped into three main classes (THC‐type, CBD‐type, and intermediate type). We tested a genetic model associating these three groups with functional and nonfunctional alleles of the cannabidiolic acid synthase gene (CBDAS).
Methods
We characterized cannabinoid content and assayed CBDAS genotypes of >300 feral C. sativa plants in Minnesota, United States. We performed a test cross to assess CBDAS inheritance. Twenty clinical cultivars obtained blindly from the National Institute on Drug Abuse and 12 Canadian‐certified grain cultivars were also examined.
Results
Frequencies of CBD‐type, intermediate‐type, and THC‐type feral plants were 0.88, 0.11, and 0.01, respectively. Although total cannabinoid content varied substantially, the three groupings were perfectly correlated with CBDAS genotypes. Genotype frequencies observed in the test cross were consistent with codominant Mendelian inheritance of the THC:CBD ratio. Despite significant mean differences in total cannabinoid content, CBDAS genotypes blindly predicted the THC:CBD ratio among clinical cultivars, and the same was true for industrial grain cultivars when plants exhibited >0.5% total cannabinoid content.
Conclusions
Our results extend the generality of the inheritance model for THC:CBD to diverse C. sativa accessions and demonstrate that CBDAS genotyping can predict the ratio in a variety of practical applications. Cannabinoid profiles and associated CBDAS segregation patterns suggest that feral C. sativa populations are potentially valuable experimental systems and sources of germplasm.
Keywords: Cannabaceae, CBDA synthase, chemotype, genetic markers, hemp, marijuana
Natural genetic variation affects the composition of phytochemicals in plants and can be of great economic importance. With a record of use by humans as food, fiber, and medicine spanning thousands of years (Faeti et al., 1996), recent decades of prohibition, and a rapidly changing regulatory context, Cannabis sativa L. (Cannabaceae) was until recently among the least studied agricultural crops (Small, 2016). Prominent among the traits of C. sativa are cannabinoids, a unique class of specialized metabolites synthesized and stored in glandular trichomes that are located on the floral bracts of pistillate inflorescences (Livingston et al., 2020; Fig. 1A–C). The ratio of the two most abundant cannabinoids, tetrahydrocannabinolic acid (THCA) and cannabidiolic acid (CBDA), hereafter THC and CBD, is represented by three main classes: THC‐type plants with THC:CBD ≥ 10, intermediate‐type plants with THC:CBD ≈ 1, and CBD‐type plants with THC:CBD ≤ 0.1 (de Meijer et al., 2003). Some authors have referred to these chemotype (chemical phenotype) classes as Type I, Type II, and Type III (Small, 2016). Here we introduce for clarity a more descriptive classification of THC‐type, intermediate‐type, and CBD‐type plants. Descriptive terms also avoid confusion associated with recent statutory definitions of C. sativa that vary widely among political jurisdictions (e.g., “industrial hemp” and “medical marijuana”). Our findings suggest that patterns of cannabinoid inheritance render some of these popular definitions inaccurate at least from a botanical perspective.
Figure 1.
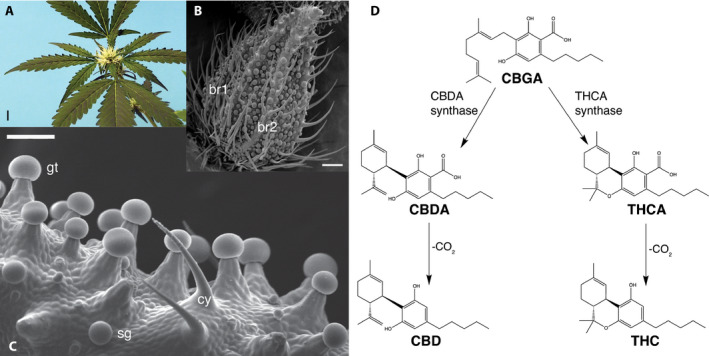
Cannabinoid biosynthesis occurs in glandular trichomes borne primarily on pistillate inflorescences composed of leaves, bracts, and florets. (A) Pistillate inflorescence with receptive, white stigmas before pollination. Scale bar = 1 cm. Photo credit: George Weiblen. (B) Scanning electron micrograph of an immature pistillate floret enclosed by a pair of imbricate bracts (br1 and br2) bearing multicellular hairs. Scale bar = 0.1 mm. Photo credit: David Marks. (C) Scanning electron micrograph of stalked glandular trichomes (gt), sessile glands (sg), and cystoliths (cy). Cystoliths are pointed hairs containing calcium carbonate crystals. Scale bar = 100 µm. Photo credit: David Marks. (D) Biosynthesis of major cannabinoid compounds. CBDA and THCA are produced enzymatically from a common CBGA precursor by different enzymes. Both compounds may be decarboxylated to pharmacologically active neutral forms CBD and THC, respectively.
Synthesis of CBD and THC involves a common precursor, cannabigerolic acid or CBGA (Taura et al., 1995) (Fellermeier et al., 2001), and the inheritance of major chemotypes is consistent with single‐locus Mendelian codominance (de Meijer et al., 2003). de Meijer et al. (2003) proposed a model in which alternate alleles determine THC‐type and CBD‐type in the respective homozygous plants, whereas heterozygous plants have the intermediate‐type. However, DNA sequencing uncovered the presence of separate but tightly linked loci for the THCA synthase (THCAS) and CBDA synthase genes (CBDAS), respectively (van Bakel et al., 2011; Onofri et al., 2015; Weiblen et al., 2015; Grassa et al., 2018 [Preprint]; Laverty et al., 2019). Given the impact of the THC:CBD ratio on various licit and illicit uses, developing molecular markers to more completely diagnose its genetic basis can aid efforts to regulate and improve C. sativa.
Predicting the THC:CBD ratio from a PCR‐based assay was first claimed by Pacifico et al. (2006), but primers were not made available. Rotherham and Harbison (2011) reported a set of single nucleotide polymorphism (SNP) markers based on differences between “drug‐type” and “fiber‐type” THCAS (Kojoma et al., 2006), but the latter THCAS sequence was reclassified by Laverty et al. (2019) as cannabichromenic acid synthase (CBCAS). These SNP markers and others derived from THCAS sequence (Staginnus et al., 2014) can distinguish THC‐type and CBD‐type plants, but they fail to differentiate THC‐type from intermediate‐type plants (Toth et al., 2020).
In an F2 mapping population derived from a cross of THC‐type and CBD‐type, we reported the correspondence of three THC:CBD ratio classes with different combinations of CBDAS alleles, noting a four‐base, frame shift deletion near the 5′ end of the CBDAS sequence in THC‐type plants (Weiblen et al., 2015). From this observation, we hypothesized a model of chemotype inheritance in which plants that are homozygous for nonfunctional and functional CBDAS have THC‐type and CBD‐type cannabinoid ratios. Plants that are heterozygous at the CBDAS locus have the intermediate type. The opportunity to test the validity of the model and its utility for predictive genotyping has only recently emerged with greater access to diverse populations of C. sativa (Toth et al., 2020).
We applied a research protocol approved by the U. S. Drug Enforcement Administration (DEA) to study feral C. sativa in the Minnesota River Valley (Fig. 2), aiming to test the CBDAS inheritance model that emerged from our previous mapping study. We confirmed that cannabinoid profiles and CBDAS genotypes of feral individuals are congruent with the model, as was the segregation of genotypes and phenotypes in a feral test cross. Additionally, we tested samples obtained blindly from the National Institute on Drug Abuse (NIDA) to validate the model by predicting the cannabinoid ratios of 20 clinical cultivars. Lastly, we demonstrated the utility of the CBDAS assay for predicting THC:CBD ratios in regulated crops by genotyping 12 Canadian‐certified grain cultivars.
Figure 2.
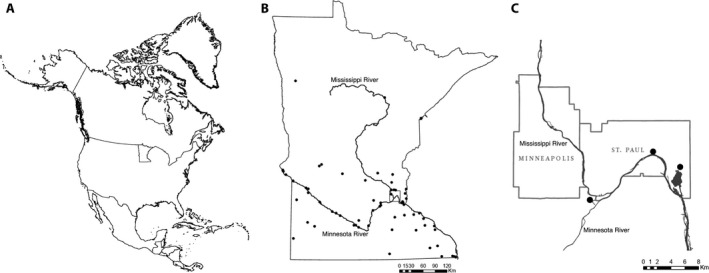
Distribution of feral Cannabis sativa in Minnesota, United States and study sample locations. (A) Location of Minnesota in North America. (B) Localities of Minnesota Department of Natural Resources observations and University of Minnesota herbarium (MIN) records from the Bell Museum. (C) Three feral population localities sampled near the confluence of the Minnesota and Mississippi rivers. Precise locations are non‐public data available upon request.
MATERIALS AND METHODS
Feral C. sativa
Minnesota feral plants were sampled in two successive years from sites located along the riparian corridor near the confluence of the Minnesota and Mississippi rivers (Fig. 2C). In 2015, we harvested all aboveground biomass of 10 mature pistillate plants from each of two locations. In 2016, we collected from the same two sites and a third location by harvesting only the terminal 15 cm of the apical infructescence of 100 mature pistillate plants each. At one of the resampled locations, three clandestinely planted individuals were encountered but excluded from statistical analysis because they did not represent the feral population. The total feral sample size was N = 317. Field harvested plants and samples were dried in the laboratory for 3 weeks at ambient conditions and separated into seed and floral fractions.
NIDA clinical C. sativa
Dried, unpollinated pistillate inflorescences representing 20 cultivars were obtained from the University of Mississippi production facility with authorization from NIDA. Cannabinoid profiles of the NIDA cultivars were not shared with the University of Minnesota until after genotyping had been completed.
Canadian industrial C. sativa
Twelve industrial cultivars were grown from Canadian‐certified planting seed during June through September 2017 at the Minnesota Agricultural Experiment Station (MAES) in Saint Paul, Minnesota. Five mature, pistillate (or monecious) plants were sampled at 70 days after planting from each cultivar, and four additional samples of each variety were collected at 105 days. Sampling of tissue at MAES was conducted in the same manner as described above for feral plants, yielding an industrial C. sativa sample of N = 108 with nine samples per cultivar.
Cannabinoid profiling
Dried floral tissue samples were analyzed using gas chromatography (ElSohly et al., 2000) to measure the percentage of total inflorescence dry mass of seven compounds: cannabichromene (CBC), CBD, cannabigerol (CBG), cannabinol (CBN), delta‐8‐tetrahydrocannabinol (d8‐THC), THC, and tetrahydrocannabivarin (THCV). We report percentage cannabinoid content as the sum of the individual compounds. Cannabinoid profiling was conducted on all field‐collected feral plants, a pistillate plant grown from feral seed in the laboratory, six test‐cross offspring of the feral parent, 20 NIDA clinical cultivars, and 108 field‐grown plants of industrial C. sativa.
CBDAS sequencing
We tested the inheritance model for the THC:CBD ratio by genotyping functional (CF) and nonfunctional alleles (CX) of CBDAS (Fig. 3A). Homozygous plants with CXCX and CFCF genotypes were predicted to have THC‐type and CBD‐type cannabinoid ratios, respectively, whereas heterozygous plants (CFCX) were predicted to have an intermediate ratio. CBDAS genotypes of 20 field‐collected feral plants, two lab‐reared feral offspring, and six test‐cross‐derived plants were determined by Sanger sequencing of ~960 bp following the method previously reported in our mapping study (Weiblen et al., 2015). PCR products were amplified with CBDAsynFor (ATG AAG TGC TCA ACA TTC) and CBDA961Rev (CCA CTC CAC CAA GGA AAA C) from gDNA isolated using a Plant DNeasy Kit (Qiagen, Hilden, Germany). Briefly, products were sequenced from the CBDAsynFor primer and aligned for comparison to reference CBDAS sequence (GenBank accession KJ469374) using Geneious 10.2.5 (Biomatters, Ltd., Auckland, New Zealand).
Figure 3.
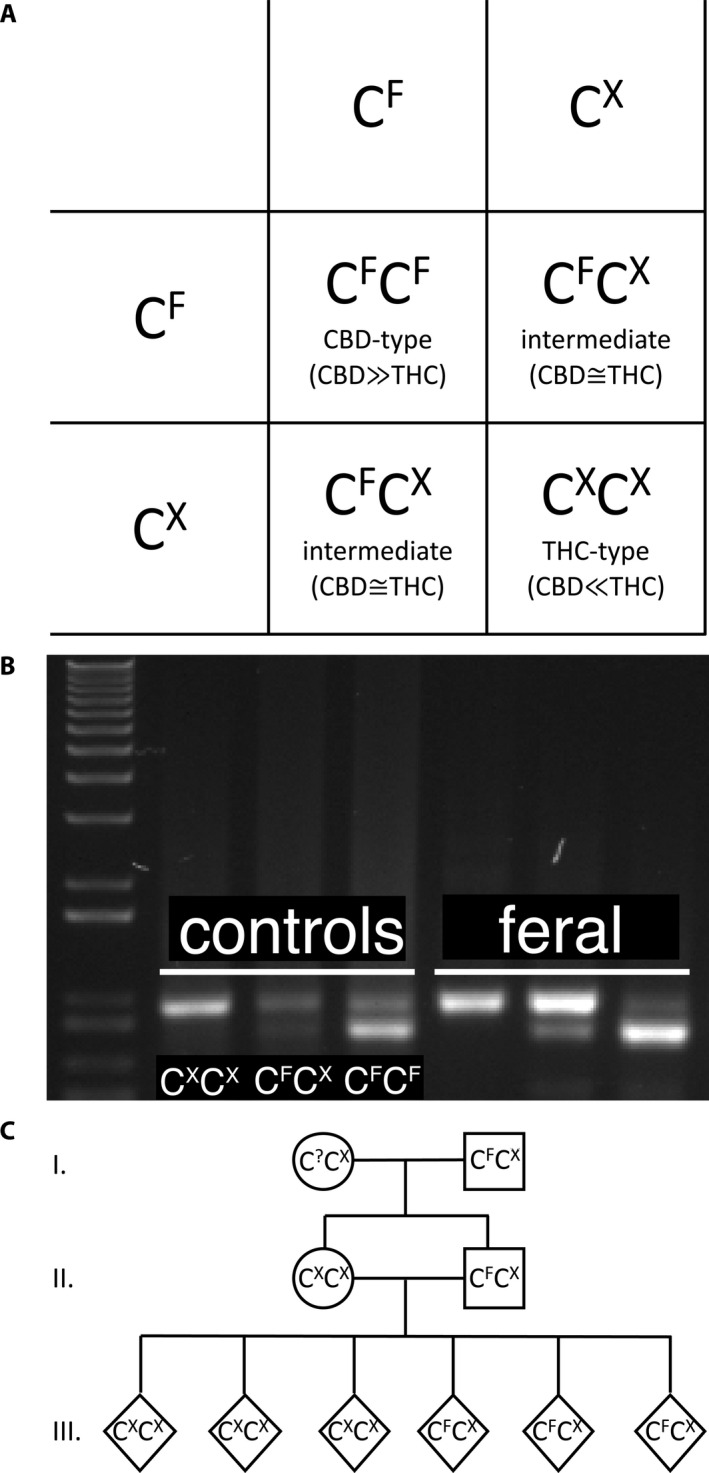
Model of major chemotype inheritance corresponding to CBDAS genotype. (A) Monohybrid inheritance in which mating between heterozygous individual yields descendants that are homozygous for functional CBDAS (CFCF), homozygous for nonfunctional CBDAS (CXCX), or heterozygous (CFCX). The three classes have predominantly CBD, predominantly THC, or intermediate chemotypes. (B) Agarose gel CAPS genotyping assay derived from presence or absence of a Bst1107I recognition site in functional (CF) and nonfunctional (CX) CBDAS alleles. In the first lane is Invitrogen 1 Kb Plus DNA Ladder (Thermo Fisher Scientific, Waltham, MA, USA). Lanes 2–4 are are verified genotypes from Weiblen et al. (2015), and lanes 5–7 are feral individuals representing each of the three genotypes. (C) Experimental verification of CBDAS inheritance by crossing a CFCX female with a CXCX male sibling offspring from a CFCX feral parent. Pedigree of (I) feral parents, (II) first generation offspring, and (III) predicted second generation offspring. The genotype of the feral pollen donor is uncertain (C?CX) but must have carried at least one non‐functional allele (CX).
CBDAS gel assay
Genotypes of 297 feral plants, 20 NIDA clinical cultivars, and 108 industrial C. sativa plants were determined using a cleaved amplified polymorphic sequence (CAPS) assay (Fig. 3B) that exploits a Bst1107I recognition site (GTATAC) present in functional (CF) but absent in nonfunctional (CX) alleles of CBDAS. PCR products for the CAPS assay were obtained as described above. DNA was isolated from floral tissue using a modified cetyl trimethylammonium bromide (CTAB) extraction buffer and organic extraction (Doyle and Doyle, 1987), precipitated using sodium acetate and ethanol, and digested with FastDigest Bst1107I (Thermo Fisher Scientific, Waltham, MA, USA). Digested fragments were separated on 0.8% agarose/1X TAE gels. Genotypes were scored by comparing digested fragment lengths of CBDAS PCR products to sequence‐verified control samples (Fig. 3B). CBDAS CAPS genotyping of 56 test cross offspring was also performed on DNA isolated from hulled seeds using the REDExtract‐N‐Amp Seed PCR Kit (Millipore Sigma, Burlington, MA, USA).
Feral offspring test cross
Using previously reported germination and growth protocols (Weiblen et al., 2015), we reared offspring of a single feral intermediate‐type plant known to be heterozygous (CFCX) for CBDAS (Fig. 3C). Among the offspring, we crossed a heterozygous pistillate plant (CFCX) with a staminate plant that was homozygous for nonfunctional CBDAS (CXCX). We collected mature seed from the cross to compare the frequency of expected and observed allelic combinations in the F2 generation using a G‐test for goodness‐of‐fit.
Statistical analyses
Multiple comparison tests (Tukey HSD) of mean cannabinoid content among feral, clinical, and industrial C. sativa plants, and among CBDAS genotypes by THC:CBD ratio classes were performed with the Least Squares function in JMP Pro 14.2.0 software (SAS, Cary, NC, USA). A likelihood ratio test for goodness‐of‐fit was calculated with the Distribution function in JMP to detect departures from codominant Mendelian inheritance. A likelihood ratio test for departure from Hardy–Weinberg equilibrium of observed feral CBDAS genotype frequencies was performed with ExactoHW 1.1 software (Engels, 2009). Cannabinoid phenotypic data and CBDAS genotypes for individual plants are provided in Appendix S1.
RESULTS
Feral cannabinoids
Among 317 pistillate, feral plants, we observed three distinct THC:CBD ratio classes as measured by gas chromatography (Table 1; Fig. 4A). Plants with a CBD‐type ratio (THC:CBD ≤ 0.1) were in greatest abundance at 88%, followed by intermediate‐type (THC:CBD ~1.0) at 11% and THC‐type (THC:CBD ≥ 10.0) at 1%. Numbers of intermediate and THC‐type plants at each of three sampling locations were 12, 20, 1, and 3, 1, 0, respectively. Total cannabinoid content per plant ranged from <1% to >10% and averaged 2.80% ± 0.09 SE (Fig. 4C). The distribution was skewed in favor of most plants (62%) having less than 3% total cannabinoid content.
Table 1.
Cannabinoid ratio groups, CBDAS genotypes, and cannabinoid content of Minnesota feral, NIDA clinical, and Canadian certified industrial C. sativa. Cannabinoid content, including total cannabinoid content (TCC) is reported as the mean percentage of inflorescence dry mass ± SE.
| N | f | Genotype | %THC (SE) | %CBD (SE) | %TCC (SE) | |
|---|---|---|---|---|---|---|
| Minnesota feral | ||||||
| CBD‐type | 280 | 0.88 | CFCF | 0.11 (0.00) | 2.40 (0.08) | 2.71 (0.09) |
| Intermediate | 33 | 0.11 | CFCX | 1.42 (0.11) | 1.85 (0.15) | 3.57 (0.28) |
| THC‐type | 4 | 0.01 | CXCX | 2.62 (0.48) | 0.21 (0.06) | 3.08 (0.53) |
| NIDA clinical | ||||||
| CBD‐type | 6 | NA | CFCF | 0.19 (0.01) | 4.94 (0.27) | 5.39 (0.30) |
| Intermediate | 11 | NA | CFCX | 3.25 (0.28) | 4.12 (0.26) | 7.91 (0.55) |
| THC‐type | 3 | NA | CXCX | 13.31 (0.27) | 0.04 (0.00) | 14.12 (0.29) |
| Industrial hemp | ||||||
| CBD‐type | 101 | 0.94 | CFCF | 0.07 (0.00) | 1.45 (0.09) | 1.62 (0.10) |
| Intermediate | 7 | 0.06 | CFCX | 0.64 (0.07) | 0.96 (0.11) | 1.71 (0.19) |
Figure 4.
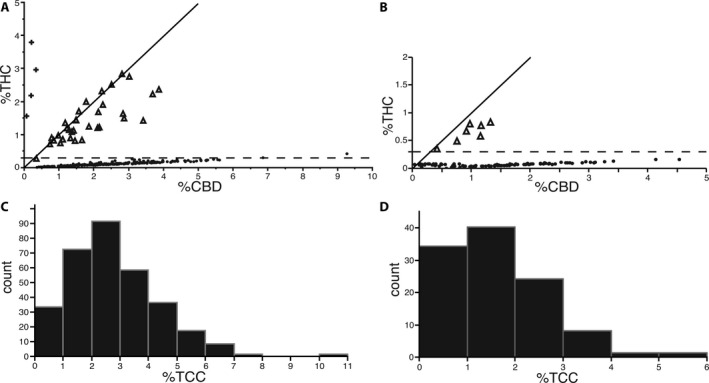
Cannabinoid profiles of Minnesota feral and industrial C. sativa sampled across 12 field‐grown Canadian certified grain and dual use cultivars. (A) Percentage dry mass THC versus CBD among 317 Minnesota feral plants. The horizontal dashed line marks the 0.3% THC statutory threshold, and the solid line indicates a 1:1 THC:CBD ratio. Symbols for CBDAS genotypes are + (CXCX), ▲ (CFCX), and ● (CFCF). (B) Percentage dry mass THC versus CBD among 108 industrial hemp plants representing 12 Canadian‐certified grain cultivars (9 samples/variety) grown in Saint Paul, Minnesota. (C) Distribution of total cannabinoid content among 317 Minnesota feral plants. (D) Distribution of total cannabinoid content among the 108 industrial hemp plants.
Feral CBDAS
CBDAS genotypes of feral plants (Table 1) matched the predictions of the model (Fig. 3A). All 280 CBD‐type plants were homozygous for functional CBDAS (CFCF), all 33 intermediate‐type plants were heterozygous (CFCX), and all four THC‐type plants were homozygous for nonfunctional CBDAS (CXCX). Allele frequencies among feral plants for functional and nonfunctional variants were CF = 0.94 and CX = 0.06, respectively. The observed frequency of the CXCX genotype (four plants) slightly exceeded the Hardy–Weinberg expectation (exact test, P = 0.0318).
Feral test cross
The test cross between a heterozygous, pistillate plant (CFCX) and a staminate plant homozygous for nonfunctional CBDAS (CXCX) was consistent with the inheritance of CBDAS genotypes according to a single‐locus Mendelian factor. Genotyping of 62 offspring of the test cross yielded nearly equal proportions of the two predicted genotypes and a few individuals of an unexpected third genotype (29 CXCX, 30 CFCX, and 3 CFCF). Genotype frequencies were not significantly different from expectations (G = 0.017; P = 0.90; Fig. 3C). The three offspring homozygous for functional CBDAS could have resulted from self‐pollination given the occurrence of occasional, adventitious staminate flowers in pistillate inflorescences. Male sex expression is commonly observed at low frequency in genetically female C. sativa plants (Hirata, 1927).
NIDA C. sativa
Blind genetic testing of 20 NIDA clinical cultivars yielded all three CBDAS genotypes (Table 1). Total cannabinoid content in the clinical samples ranged from 4.5% to 14.7% (mean = 8.1% ± 0.7 SE) and was significantly greater on average than Minnesota feral C. sativa (Tukey HSD: t = −15.08, df = 442; p < 0.0001). Regardless of total cannabinoid content, the THC:CBD ratios revealed after genotyping perfectly matched model predictions based on the observed CBDAS genotypes (Table 1).
Industrial C. sativa
Among 108 field‐grown plants representing 12 Canadian‐certified grain cultivars (including four dual use, grain/fiber cultivars), 101 individuals were homozygous for functional CBDAS (CFCF), and seven were heterozygous (CFCX). Each of the seven heterozygotes had an intermediate THC:CBD ratio and exceeded the <0.3% THC statutory threshold (Table 1; Fig. 4B). A subset of the homozygous plants (20 of 101) had intermediate THC:CBD ratios but had significantly lower total cannabinoid content (mean = 0.40% ± 0.04 SE) than either heterozygous intermediates (mean = 1.71% ± 0.19 SE; Tukey HSD: t = 3.62, df = 105, P = 0.0013) or homozygous CBD‐type plants (mean = 1.93% ± 0.10 SE; Tukey HSD: t = 7.42, df = 105, P < 0.0001). Compared to feral C. sativa, Canadian‐certified cultivars collectively had significantly lower cannabinoid content (mean = 1.63% ± 0.10 SE; Tukey HSD: t = 6.93, df = 442, P < 0.0001) and a similarly skewed distribution but with a far greater proportion of plants (90%) containing less than 3.0% total cannabinoid content (Fig. 4D).
DISCUSSION
Cannabinoids
It is not widely known that the discovery of CBD can be traced to Minnesota where it was first isolated from fiber‐type C. sativa (Adams et al., 1940). Loss of access to Philippine fiber during World War II prompted the Defense Plant Corporation, a U. S. federal agency, to temporarily license and subsidize C. sativa cultivation in Minnesota (Dewey, 1943). Escape and naturalization resulted in widespread populations of feral C. sativa throughout the upper Midwest (Nugent, 1938; Schoenrock, 1966). We used Minnesota feral C. sativa to validate a predictive model for the inheritance of the THC:CBD ratio. Genotyping of clinical and industrial C. sativa suggests a general association between allelic variation at the CBDAS locus and the three main classes of the THC:CBD ratio.
CBDAS in feral C. sativa
The complete correspondence of THC:CBD ratio classes with CBDAS genotypes in Minnesota feral C. sativa provides strong evidence for the generality of the model of inheritance that was derived from inbred biparental genetic mapping (Fig. 3A) (Weiblen et al., 2015). Also supporting generality were sequences from 20 feral, THC‐type or intermediate plants sharing the same diagnostic four‐base frameshift deletion previously reported for nonfunctional CBDAS isolated from THC‐type C. sativa (Weiblen et al., 2015). Using the CAPS assay based on this diagnostic deletion, an additional 297 feral plants also associated CBDAS genotypes with the three THC:CBD ratio groups. The marginally significant departure of CBDAS genotype frequencies from Hardy–Weinberg equilibrium might represent a Type I statistical error given only four THC‐type plants among 317 individuals. Additional study is needed to evaluate the potential for non‐equilibrium processes to have shaped feral CBDAS genotype frequencies, including the possibility of phenotypic selection for cannabinoids as deterrents to seed predation (Small, 2016).
Identifying THC‐type plants among first generation offspring of a feral intermediate plant signaled the presence of pollen donors harboring THC‐type alleles as would be expected in a segregating field population. Experimental crossing of offspring from a feral intermediate produced 59 of 62 genotypes that were consistent with the expected segregation of CBDAS genotypes in the subsequent generation.
Unexplained cannabinoid variation
Despite evidence from feral C. sativa for incomplete dominance affecting the THC:CBD ratio, variance within each of the three ratio classes remains unexplained and deserves further study. Similar to the segregating population of Weiblen et al. (2015), variation was greatest among the feral intermediate class (Fig. 4A), and intermediate‐type plants had slightly higher quantities of CBD than THC. Considering that these two cannabinoids share a common precursor (CBG), we speculate that CBDAS is a superior competitor to THCAS either in terms of substrate affinity or catalytic efficiency. Industrial C. sativa had the same pattern (Fig. 4B), although with less variation within classes than feral C. sativa, as would be expected for a domestic crop variety.
Another major component of cannabinoid variation not explained by the THC:CBD ratio model is the nearly 10‐fold range in total cannabinoid content (Fig. 4C, D). A broad range of variation was observed in all three ratio classes (Fig. 4A, B), indicating that the inheritance of cannabinoid quantity is independent of the THC:CBD ratio. Quantitative trait locus (QTL) analysis identified QTL for total cannabinoid content on five different chromosomes (Grassa et al., 2018 [Preprint]), and none were linked to the single QTL that is strongly associated with the THC:CBD ratio or the genes for CBDAS and THCAS on chromosome 7 (Fig. 5A). Whether any of the independent QTLs are associated with differential gene regulation affecting CBD or THC production is an intriguing question for future study. Alternatively, it has been hypothesized that cannabinoid gene copy number influences potency (Kovalchuk et al., 2020).
Figure 5.
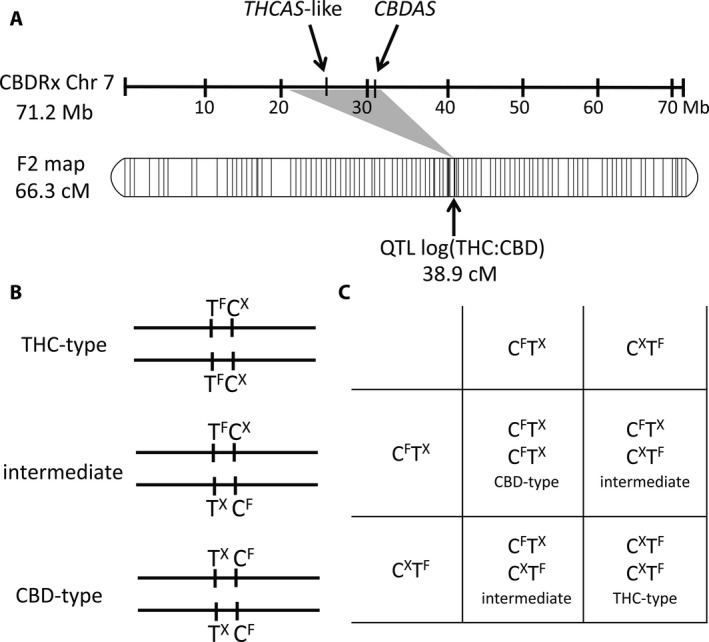
(A) Alignment of a chromosome 7 physical map for C. sativa (Grassa et al., 2018) with the corresponding linkage group from the segregating F2 population of Weiblen et al. (2015). A single QTL at 38.9 cM, accounting for >90% of variance in the THC:CBD ratio, maps to a region between 20.6 Mb and 31.5 Mb on chromosome 7. Genes for THCAS and CBDAS are located within this region at 26 Mb and 31 Mb, respectively. (B) Model in which THC‐type and CBD‐type plants are reciprocally homozygous for alternately functional (f) and nonfunctional (x) alleles of THCAS and CBDAS. Intermediate‐type plants are heterozygous at both loci. (C) Punnett square for selfing the F1 intermediate of a hybrid cross. According to the close linkage of the loci, we predict that recombinants are not likely to be observed among the F2 descendants of hybrid crosses.
Assaying clinical and industrial C. sativa
We extended the utility of the CBDAS assay by blindly genotyping 20 clinical cultivars obtained from the NIDA production facility in Mississippi. Only after genotyping were the cannabinoid profiles from Mississippi shared with the investigators in Minnesota. CBDAS genotypes predicted the three THC:CBD ratio classes with 100% accuracy, suggesting that CBDAS genotypes can generally predict THC:CBD ratios in drug cultivars. Applying the assay to a set of 108 samples from 12 Canadian‐certified industrial cultivars (grain and dual‐use) also suggest broad diagnostic utility.
We were surprised to discover that 6% of plants grown from Canadian‐certified seed exceeded the <0.3% THC statutory definition of industrial C. sativa. All seven plants had an intermediate THC:CBD ratio and the expected heterozygous CBDAS genotype. Had THC testing been performed on homogenized samples taken from multiple plants or if the results of several independent tests were averaged for a given population, the low frequency of noncompliant plants could have escaped detection during the multiplication of certified seed. Even the most stringent THC testing when developing foundation seed or multiplying certified seed could be confounded by pollen drift from nearby feral or cultivated C. sativa harboring THC‐type alleles (Small and Antle, 2003). Stokes et al. (2000) reported up to 36% C. sativa pollen during August in airborne pollen samples from the midwestern United States. Aerodynamic models of pollen dispersal could be tested in the field to evaluate whether the recommended isolation distance of 5 km is sufficient to maintain the genetic purity of a C. sativa crop. Regardless, the contractual obligation to neither plant nor multiply seed saved from a certified crop is especially important given the potential for pollen contamination to increase the frequency of plants with >0.3% THC in subsequent generations.
There are several situations in which the CBDAS genotype assay might have practical value. Many populations of C. sativa classified as "hemp" are segregating for THC‐type alleles (Toth et al., 2020). The risk of THC‐type alleles contaminating certified fiber and grain cultivars could be minimized by screening seed lots at all stages of breeding and multiplication to assure purity. When seed provenance is uncertain or questionable, genetic testing of samples prior to planting could reduce the risk of non‐compliant crops. Farming “industrial hemp” is a risky proposition at present because crops are tested near maturity for THC regulatory compliance.
The homozygous functional CBDAS genotype accurately identified all 101 industrial C. sativa plants that were compliant with the <0.3% THC statutory definition of industrial hemp. However, 20 plants among the compliant individuals had exceptionally low total cannabinoid content (<0.5%). When cannabinoids were this scarce, the THC:CBD ratio was closer to intermediate type than to either the THC‐type or CBD‐type. Uncertainty associated with sample preparation or gas chromatography in the range of ±0.1–0.3% might have skewed the THC:CBD ratio and obscured the distinction between the intermediate and CBD‐type. Alternatively, novel genetic mechanisms might await discovery. In any event, the homozygous functional CBDAS genotype correctly predicted that even these 20 plants were compliant with the <0.3% THC statutory definition of industrial C. sativa.
THC:CBD ratio inheritance model
The CBDAS genotype accurately predicted all three THC:CBD ratio classes in diverse C. sativa gene pools, supporting its broad utility. The simplest genetic explanation for this association would be if CBDAS and THCAS represent alternate alleles of a single locus (de Meijer et al., 2003). However, there are at least two or more tightly linked cannabinoid synthase loci in diverse accessions of C. sativa (van Bakel et al., 2011; Onofri et al., 2015; Weiblen et al., 2015; Grassa et al., 2018 [Preprint]; Laverty et al., 2019). Recent annotation of several C. sativa genome assemblies (Grassa et al., 2018 [Preprint]; Laverty et al., 2019), including one anchored with whole‐genome shotgun sequencing of an F2 mapping population (Weiblen et al., 2015), indicate that separate loci for THCAS and CBDAS are tightly linked in a chromosomal region of highly suppressed recombination. Grassa et al. (2018; Preprint) proposed the physical arrangement of the synthases in close proximity on chromosome 7 (Fig. 5A). Based on these findings, we suggest a mechanistic model explaining the predictive power of the CBDAS genotype with respect to THC:CBD inheritance. In CBD‐type plants, the expressed CBDAS is tightly linked to a nonfunctional and/or under‐expressed THCAS. In THC‐type plants, a nonfunctional and underexpressed CBDAS is tightly linked to a functional and overexpressed THCAS. Tight genetic linkage of the synthases renders CBD‐type and THC‐type plants reciprocally homozygous for the respective alleles, whereas intermediate plants are reciprocally heterozygous.
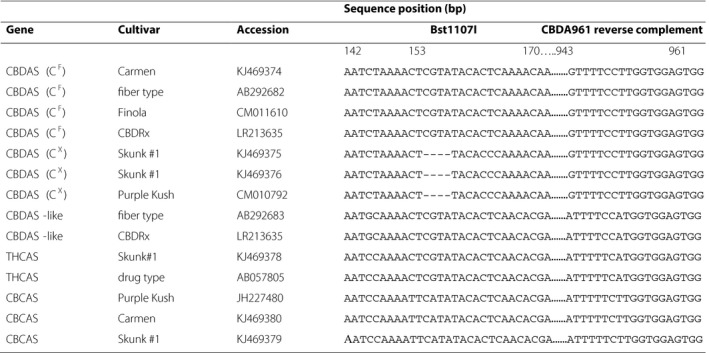
The conserved four‐base deletion at position 153 in the CBDAS sequence that renders it nonfunctional (Weiblen et al., 2015) and is the target of the CAPS assay appears to exist in THC‐type and intermediate plants among diverse C. sativa accessions (Table 2). Although the model of incomplete dominance for the THC:CBD ratio seems robust, it does not completely account for cannabinoid inheritance. In particular, the potential influence of gene copy number on cannabinoid expression deserves further consideration (Kovalchuk et al., 2020).
Table 2.
Cannabinoid synthase sequence alignment including the four‐base deletion that distinguishes functional (CF) from nonfunctional (CX) alleles of cannabidiolic acid synthase (CBDAS). A Bst1107I restriction site spans the deletion. Downstream sequences from 943 to 961 bp illustrate how the reverse primer CBDA961Rev selectively amplifies CBDAS over other cannabinoid synthases. Gene sequences are annotated as CBDAS, CBDAS‐like, THCAS, or CBCAS according to Grassa et al. (2018) for cultivars including Carmen hemp (Weiblen et al., 2015), CBDRx (Grassa et al., 2018), drug type (Sirikantaramas et al., 2004), fiber type (Taura et al., 2007), Finola (Laverty et al., 2019), Purple Kush (Laverty et al., 2019), and Skunk #1 (Weiblen et al., 2015).
CBDAS versus THCAS
When the first CBDAS sequence was published Taura et al. (2007) reported additional homologs in fiber‐type C. sativa. Subsequent studies have suggested that cannabinoid synthase and synthase‐like sequences are likely duplicated (Onofri et al., 2015; Weiblen et al., 2015). BLAST searches with the original CBDAS sequence return no fewer than 37 hits described as CBDAS compared to 94 hits described as THCAS with the first reported THCAS sequence (Sirikantaramas et al., 2004). The CBDA961Rev primer reported here amplifies CBDAS (CF) and nonfunctional CBDAS (CX) to the exclusion of potentially confounding cannabinoid synthase gene copies (Table 2). The association of functional and nonfunctional CBDAS alleles with THC‐type plants and intermediate plants has also been independently affirmed in a broader survey of European C. sativa (Cascini, 2019) and in feral New York populations (Toth et al., 2020). Functional CBDAS is present in the genome of the CBD‐type cultivar Finola but absent from the THC‐type Purple Kush (Laverty et al., 2019). As predicted by the model, a CBDAS sequence bearing the four‐base (CX) deletion is found in the Purple Kush genome assembly but not in Finola.
We suggest that CBDAS genotype assays may be more informative than existing THCAS polymorphisms because the latter alone do not yet distinguish THC‐type plants from intermediates (Rotherham and Harbison, 2011; Staginnus et al., 2014). One explanation for the limited diagnostic power of THCAS‐based assays is that a conserved and presumably nonfunctional allele of THCAS has not been identified. Cannabinoid synthase sequences from both drug and fiber cultivars previously thought to be copies of THCAS (Kojoma et al., 2006; Weiblen et al., 2015) turned out to be cannabichromenic acid synthase (CBCAS) according to Laverty et al. (2019). Adding to the confusion, sequences from CBD‐type plants annotated by Rotherham and Harbison (2011) as THCAS are more similar to CBCAS (Grassa et al., 2018 [Preprint]; Laverty et al., 2019). It is possible that the high sequence similarity of different synthase genes and subsequent misidentification have confounded efforts to develop diagnostic assays of the THC:CBD ratio based on THCAS polymorphisms. Perhaps it is the highly suppressed recombination in the physical region harboring reciprocally linked, functional and nonfunctional alleles that maintains the single locus‐like pattern of cosegregation. This hypothesis predicts the existence of a nonfunctional THCAS allele that might yield a THCAS‐based codominant assay similar to the CBDAS‐based assay (Fig. 5C).
Feral problems and prospects
Due to research restrictions resulting from the Controlled Substances Act of 1970, few data on the cannabinoid profiles or genetics of feral C. sativa were available until recently (Datwyler and Weiblen, 2006). The rarity of THC‐type plants in feral populations and the odds of locating one‐in‐a‐hundred are consistent with the popular opinion that “ditch weed” is non‐intoxicating. Our findings are at the same time consistent with law enforcement concerns about feral populations concealing drug‐type C. sativa, although their low frequency undercuts the efficiency of drug eradication. We would like to know whether the presence of THC‐type and intermediate‐type plants in feral populations is due to the impurity of historical fiber cultivars or to more recent introgression of THC‐type genetics from clandestine drug operations.
Two arguments favor historical impurity. First, THC and CBD were not characterized chemically until long after the introduction of fiber hemp to Minnesota such that neither breeders nor producers of hemp could accurately assess purity. Second, THC‐type pollen is unlikely to be abundant in the landscape relative to feral pollen based on horticultural practice for drug production. Unpollinated inflorescences are known to invest resources in excessive trichome formation and cannabinoid production (Merlin and Clark, 2013). Drug operations either cultivate clonally propagated pistillate plants (females) or remove staminate plants (males) to avoid pollination and increase the potency of the crop.
Alternatively, the nonfunctional CBDAS allele might have been introduced to feral populations by dispersal of THC‐type pollen from cultivation and escape of drug‐type C. sativa. Pollen can travel distances up to kilometers in wind‐pollinated species such as C. sativa, and long‐distance seed dispersal by birds has even greater potential to influence the migration of genes among populations (Small, 2016). Additional work is needed to examine population genetic patterns and processes by sampling feral C. sativa geographically and by comparing locations with different histories of fiber and drug cultivation.
Feral populations could be a valuable source of germplasm for breeding and provide opportunities to investigate the inheritance of quantitative variation in total cannabinoid content (potency). Genetic mechanisms influencing cannabinoid quantity are currently a subject of untested speculation. None of the QTL for potency that have been identified are linked to THCAS or CBDAS but are rather associated with different chromosomes (Weiblen et al., 2015; Grassa et al., 2018 [Preprint]). Breeding to increase the yield of CBD while maintaining THC levels below the legal limit is an obvious potential application, as is breeding for fiber and grain yield. At least 70 generations of feral adaptation to regional soil and climate conditions present both opportunities and challenges for re‐domestication.
Broader implications
The presence of more than one of the three cannabinoid classes in feral, industrial, and clinical populations renders the dichotomy between “hemp” and “marijuana” meaningless from a botanical perspective. Other classes of plants add to the complexity of C. sativa. For example, Fournier (2004) reported cultivars producing mostly CBG, the common precursor to THC and CBD. We avoided using common names not simply because they misrepresent patterns in nature and fail to match statutory intent. The dichotomy between “hemp” and “marijuana” perpetuates culturally biased and pejorative assumptions about C. sativa that have hindered scientific investigation for nearly a century (Abel, 1980). Rooted in colonial history, North American C. sativa has literally and figuratively escaped from that past, adapting and thriving today across a diverse array of natural ecosystems and engineered systems. A decolonized definition recognizing THC‐type, CBD‐type, intermediate‐type, and CBG‐type plants would be more accurate botanically and perhaps more practical as the use and regulation of C. sativa continues to expand and diversify.
AUTHOR CONTRIBUTIONS
J.P.W., C.J.D., and G.D.W. conceived of the study and wrote the manuscript. J.P.W., C.J.D., and G.D.W. collected the feral and industrial C. sativa. J.P.W. and C.J.D. performed the genetic analyses and conducted the test cross experiment. M.A.E., S.C., M.M.R., and C.G.M. cultivated and provided clinical samples and performed the chemical analyses. All authors participated in review and revision of the final manuscript.
Supporting information
APPENDIX S1. Cannabinoid phenotypic data and CBDA synthase genotypes.
Acknowledgments
This study was supported by the Minnesota Department of Agriculture (H006142601 & H007044801) and Dr. Bronner’s Magic Soaps (All One God Faith, Inc.). We acknowledge the National Institute on Drug Abuse, Contract #N01DA‐15‐7793 for providing the clinical samples and for providing analytical support (cannabinoids analysis of all samples) for this work. We thank Dr. Bronner’s Cosmic Engagement Officer, David Bronner, for patience and John Partridge, U. S. Drug Enforcement Administration, for diligence in developing a protocol for field collection of feral C. sativa. We acknowledge permission from the Minnesota Department of Natural Resources, the Minnesota Department of Transportation, and Saint Paul Parks and Recreation for access to field locations. We thank M. David Marks and the University of Minnesota College of Biological Sciences Imaging Center for assistance with microscopy and Veronica Tonnell for laboratory assistance. Margaret Wiatrowski, Anthony Cortilet, and Denise Thiede facilitated our participation in the Minnesota Industrial Hemp Pilot Program and with the importation of Canadian‐certified industrial C. sativa. We are grateful to the offices of the General Counsel and the Vice President for Research at the University of Minnesota, the U. S. congressional delegation for Minnesota, the Minnesota Board of Pharmacy, and the U. S. Drug Enforcement Administration for the opportunity to do this study. We also thank the reviewers of the manuscript for critical insights that helped us to improve the presentation.
Wenger, J. P. , Dabney C. J. III, ElSohly M. A., Chandra S., Radwan M. M., Majumdar C. G., and Weiblen G. D.. 2020. Validating a predictive model of cannabinoid inheritance with feral, clinical, and industrial Cannabis sativa . American Journal of Botany. 107(10): 1423–1432.
Data Availability
Additional Supporting Information may be found online in Appendix S1.
LITERATURE CITED
- Abel, E. L. 1980. Marihuana: The first twelve thousand years. Plenum Press, New York, NY, USA. [Google Scholar]
- Adams, R. , Pease D. C., and Clark J. H.. 1940. Isolation of cannabinol, cannabidiol and quebrachitol from red oil of Minnesota wild hemp. Journal of the American Chemical Society 62: 2194–2200. [Google Scholar]
- Cascini, F. , Farcomeni A., Migliorini D., Baldassarri L., Boschi I., Martello S., Amaducci S., Lucini L., and Bernardi J.. 2019. Highly predictive genetic markers distinguish drug‐type from fiber‐type Cannabis sativa L. Plants 8: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datwyler, S. L. , and Weiblen G. D.. 2006. Genetic variation in hemp and marijuana (Cannabis sativa L.) according to amplified fragment length polymorphisms. Journal of Forensic Sciences 51: 371–375. [DOI] [PubMed] [Google Scholar]
- de Meijer, E. P. M. , Bagatta M., Carboni A., Crucitti P., Moliterni V. M. C., Ranalli P., and Mandolino G.. 2003. The inheritance of chemical phenotype in Cannabis sativa L. Genetics 163: 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey, L. H. 1943. Fiber production in the Western Hemisphere. Miscellaneous Publication no. 518, 63–69. U.S. Department of Agriculture, U. S. Government Printing Office, Washington, D.C., USA. [Google Scholar]
- ElSohly, M. A. , Ross S. A., Mehmedic Z., Arafat R., Yi B., and Banahan B. F.. 2000. Potency trends of delta9‐THC and other cannabinoids in confiscated marijuana from 1980‐1997. Journal of Forensic Sciences 45: 24–30. [PubMed] [Google Scholar]
- Engels, W. R. 2009. Exact tests for Hardy‐Weinberg proportions. Genetics 183: 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faeti, V. , Mandolino G., and Ranalli P.. 1996. Genetic diversity of Cannabis sativa germplasm based on RAPD markers. Plant Breeding 115: 367–370. [Google Scholar]
- Fellermeier, M. , Eisenreich W., Bacherr A., and Zenk M. H.. 2001. Biosynthesis of cannabinoids: incorporation experiments with 13C‐labeled glucoses. European Journal of Biochemistry 268: 1596–1604. [DOI] [PubMed] [Google Scholar]
- Fournier, G. , Beherec O., and Bertucelli S.. 2004. Santhica 23 et 27: deux variétés de chanvre (Cannabis sativa L.) sans ∆‐9‐THC. Annales de Toxicologie Analytique XVI: 128–132. [Google Scholar]
- Grassa, C. J. , Wenger J. P., Dabney C., Poplawski S. G., Motley T., Michael T. P., Schwartz C., and Weiblen G. D.. 2018. A complete Cannabis chromosome assembly and adaptive admixture for elevated cannabidiol (CBD) content. bioRxiv 458083 [Preprint]. [Google Scholar]
- Hirata, K. 1927. Sex determination in hemp (Cannabis sativa L.). Journal of Genetics 19: 65–79. [Google Scholar]
- Kojoma, M. , Seki H., Yoshida S., and Muranaka T.. 2006. DNA polymorphisms in the tetrahydrocannabinolic acid (THCA) synthase gene in “drug‐type” and “fiber‐type” Cannabis sativa L. Forensic Science International 159: 132–140. [DOI] [PubMed] [Google Scholar]
- Kovalchuk, I. , Pellino M., Rigault P., van Velzen R., Ebersbach J., Mau R. A. J. M., et al. 2020. The genomics of Cannabis and its close relatives. Annual Review of Plant Biology 71: 713–739. [DOI] [PubMed] [Google Scholar]
- Laverty, K. U. , Stout J. M., Sullivan M. J., Shah H., Gill N., Holbrook L., Deikus G., et al. 2019. A physical and genetic map of Cannabis sativa identifies extensive rearrangements at the THC/CBD acid synthase loci. Genome Research 29: 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston, S. J. , Quilichini T. D., Booth J. K., Wong D. C. J., Rensing K. H., Laflamme‐Yonkman J., Castellarin S. D., et al. 2020. Cannabis glandular trichomes alter morphology and metabolite content during flower maturation. Plant Journal 101: 37–56. [DOI] [PubMed] [Google Scholar]
- Merlin, M. D. , and Clark R. C.. 2013. Cannabis: evolution and ethnobotany. University of California Press, Berkeley, CA, USA. [Google Scholar]
- Nugent, H. T. 1938. Report of survey: commercialized hemp (1934–35 crop) in the state of Minnesota [correspondence with H. J. Anslinger]. Treasury Department, Bureau of Narcotics, Minneapolis, MN, USA. [Google Scholar]
- Onofri, C. , de Meijer E. P. M., and Mandolino G.. 2015. Sequence heterogeneity of cannabidiolic‐ and tetrahydrocannabinolic acid‐synthase in Cannabis sativa L. and its relationship with chemical phenotype. Phytochemistry 116: 57–68. [DOI] [PubMed] [Google Scholar]
- Pacifico, D. , Miselli F., Micheler M., Carboni A., Ranalli P., and Mandolino G.. 2006. Genetics and marker‐assisted selection of the chemotype in Cannabis sativa L. Molecular Breeding 17: 257–268. [Google Scholar]
- Rotherham, D. , and Harbison S. A.. 2011. Differentiation of drug and non‐drug Cannabis using a single nucleotide polymorphism (SNP) assay. Forensic Science International 207: 193–197. [DOI] [PubMed] [Google Scholar]
- Schoenrock, R. E. 1966. Hemp in Minnesota during the wartime emergency. M.Sc. thesis, Mankato State College, Mankato, MN, USA. [Google Scholar]
- Sirikantaramas, S. , Morimoto S., Shoyama Y., Ishikawa Y., Wada Y., Shoyama Y., and Taura F.. 2004. The gene controlling marijuana psychoactivity: molecular cloning and heterologous expression of delta1‐tetrahydrocannabinolic acid synthase from Cannabis sativa L. Journal of Biological Chemistry 279: 39767–39774. [DOI] [PubMed] [Google Scholar]
- Small, E. 2016. Cannabis: a complete guide. CRC Press, Boca Raton, FL, USA. [Google Scholar]
- Small, E. , and Antle T.. 2003. A preliminary study of pollen dispersal in Cannabis sativa in relation to wind direction. Journal of Industrial Hemp 8: 37–50. [Google Scholar]
- Staginnus, C. , Zorntlein S., and de Meijer E.. 2014. A PCR marker linked to a THCA synthase polymorphism is a reliable tool to discriminate potentially THC‐rich plants of Cannabis sativa L. Journal of Forensic Sciences 59: 919–926. [DOI] [PubMed] [Google Scholar]
- Stokes, J. R. , Hartel R., Ford L. B., and Casale T. B.. 2000. Cannabis (hemp) positive skin tests and respiratory symptoms. Annals of Allergy, Asthma & Immunology 85: 238–240. [DOI] [PubMed] [Google Scholar]
- Taura, F. , Morimoto S., and Shoyama Y.. 1995. Cannabinerolic acid, a cannabinoid from Cannabis sativa . Phytochemistry 39: 457–458. [Google Scholar]
- Taura, F. , Sirikantaramas S., Shoyama Y., Yoshikai K., Shoyama Y., and Morimoto S.. 2007. Cannabidiolic‐acid synthase, the chemotype‐determining enzyme in the fiber‐type Cannabis sativa . FEBS Letters 581: 2929–2934. [DOI] [PubMed] [Google Scholar]
- Toth, J. A. , Stack G. M., Cala A. R., Carlson C. H., Wilk R. L., Crawford J. L., Viands D. R., et al. 2020. Development and validation of genetic markers for sex and cannabinoid chemotype in Cannabis sativa L. Global Change Biology Bioenergy 12: 213–222. [Google Scholar]
- van Bakel, H. , Stout J. M., Cote A. G., Tallon C. M., Sharpe A. G., Hughes T. R., and Page J. E.. 2011. The draft genome and transcriptome of Cannabis sativa . Genome Biology 12: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiblen, G. D. , Wenger J. P., Craft K. J., ElSohly M. A., Mehmedic Z., Treiber E. L., and Marks M. D.. 2015. Gene duplication and divergence affecting drug content in Cannabis sativa . New Phytologist 208: 1241–1250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Cannabinoid phenotypic data and CBDA synthase genotypes.
Data Availability Statement
Additional Supporting Information may be found online in Appendix S1.


