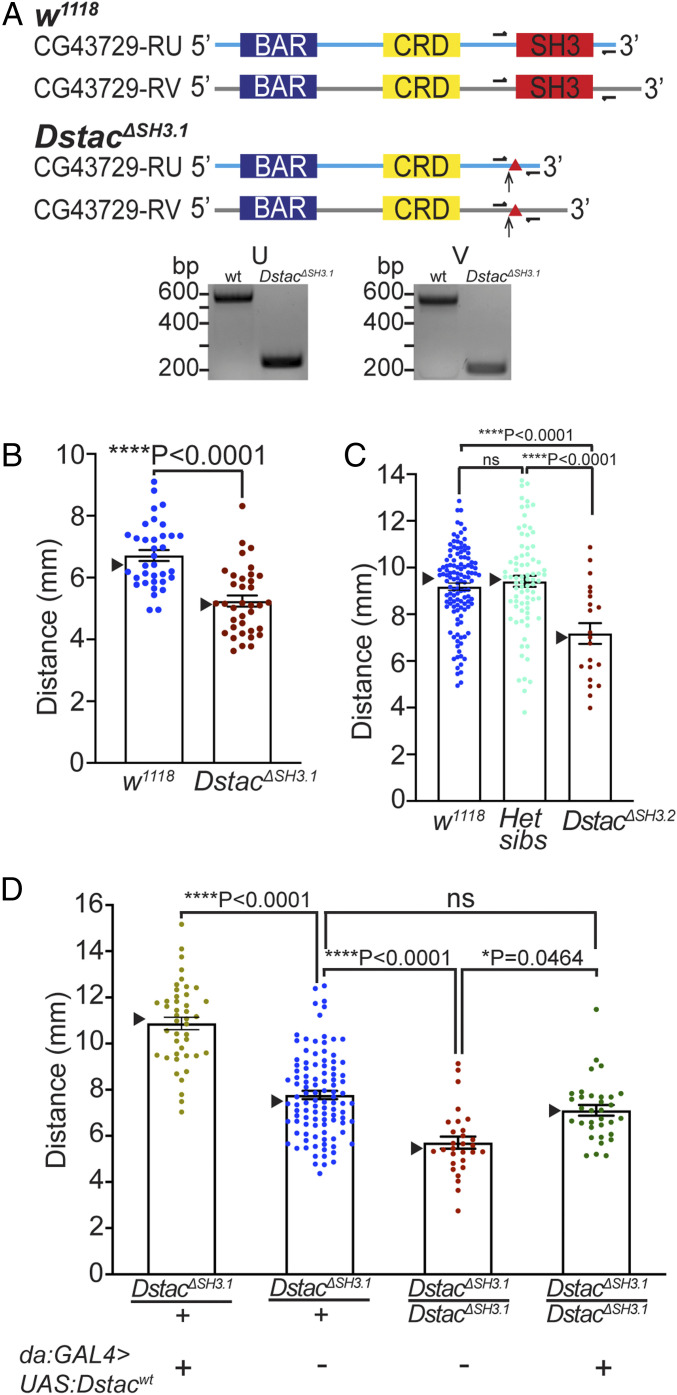
Fig. 2.
Dstac mutants generated by CRISPR-Cas9 showed reduced locomotion. (A) Schematic of two Dstac protein variants (CG43729-RU and CG43729-RV) is shown. Arrows denote the location where early stop codons occurred due to the indels (triangles) created by CRISPR-Cas9. Half arrows denote primer sites used to sequence the mutations. All primers used are listed in SI Appendix, Table S1 and Supplementary Materials and Methods. The cDNA PCR fragments of DstacΔSH3.1 are smaller than wt for both RU and RV variants, suggesting the deletions of the SH3 domain in the Dstac cDNA. The deletions of SH3 were confirmed by sequencing the wt and DstacΔSH3.1 cDNA bands (SI Appendix, Supplementary Text). The translated mutant cDNA sequences showed that an early stop codon was produced in DstacΔSH3.1 (SI Appendix, Supplementary Text). The schematic is not to scale. (B) Offsprings of founder 1 after five generations of outcrossing (DstacΔSH3.1) showed decreased locomotion compared with wt (wt, n = 35; DstacΔSH3.1, n = 36; one-tailed, unpaired t test). In this histogram and all subsequent ones, SEMs are shown, and triangles denote the median. (C) Offsprings of founder 2 after 10 generations of outcrossing (DstacΔSH3.2, n = 21) showed decreased locomotion compared with wt (n = 120) and heterozygous siblings (n = 78) (one-way ANOVA, Tukey’s multiple comparisons test). ns denotes not significant. (D) Expression of Dstacwt by heterozygous DstacΔSH3.1 larvae (n = 43) increased locomotion compared with heterozygous controls (n = 104), and expression of Dstacwt by homozygous DstacΔSH3.1 (n = 33) rescued DstacΔSH3.1 (n = 29) locomotion (Kruskal–Wallis test, Dunn’s multiple comparisons test). The larvae were genotyped by PCR after the locomotion assay.