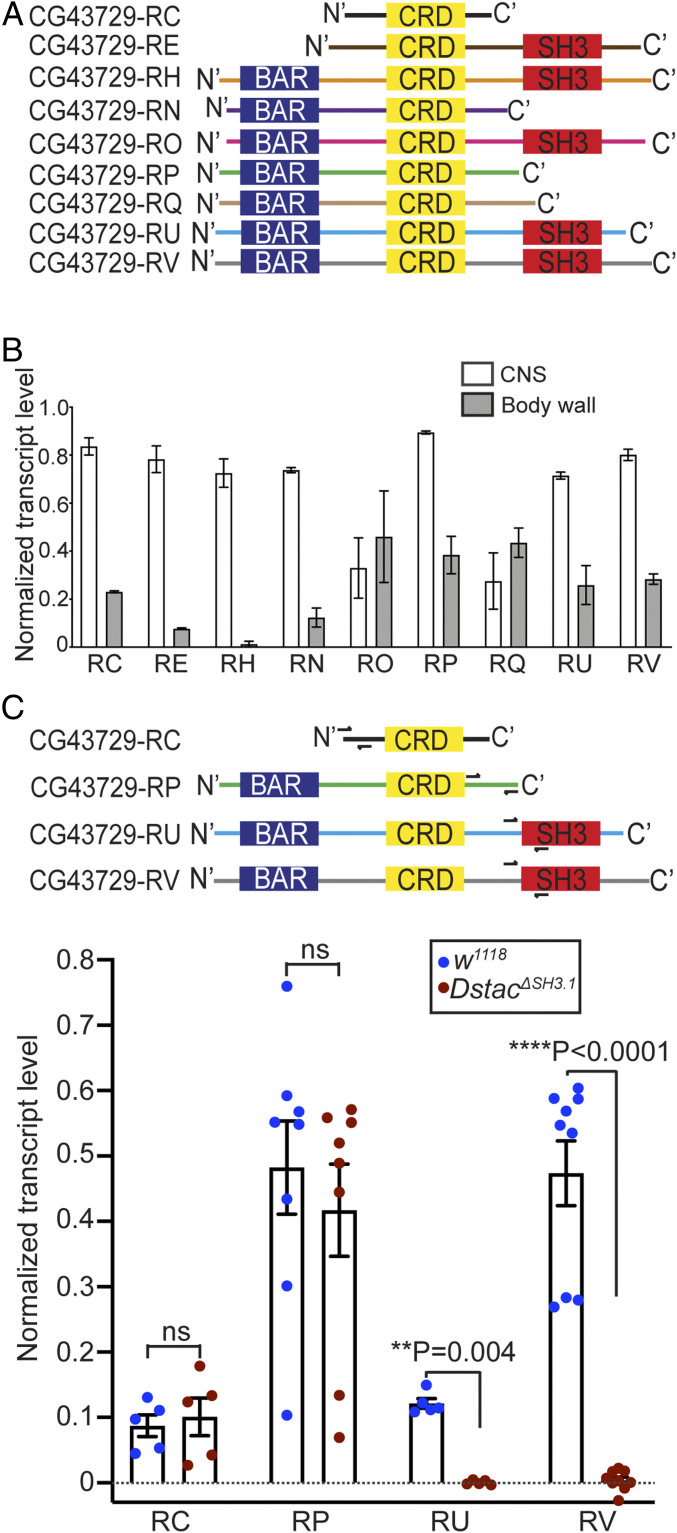
Fig. 3.
Dstac isoforms that contain the SH3 domain are affected in DstacΔSH3.1. (A) Schematic of domain composition of nine Dstac protein variants. Not all variants contain an SH3 domain. (B) Quantification of Dstac transcript levels in wt third instar larval CNS and body wall assayed by RT-PCR. Error bars represent SEM. RT-PCR primers for Dstac (CG43729) transcripts and GAPDH used for normalization can be found in SI Appendix, Table S1 and Supplementary Materials and Methods. (C) Two isoforms without the SH3 domain (CG43729-RC and CG43729-RP) and two isoforms that contain the SH3 domain (CG43729-RU and CG43729-RV) were PCR amplified from wt and DstacΔSH3.1 cDNA. Half arrows denote the primer sites. The levels of isoforms that don’t have SH3 domain (RC and RP) were comparable between wt and DstacΔSH3.1 (Mann–Whitney U test), but the isoforms that normally contained the SH3 domain (RU and RV) were diminished in DstacΔSH3.1 (Mann–Whitney U test) (RC: wt n = 5, DstacΔSH3.1 n = 5; RP: wt n = 8, DstacΔSH3.1 n = 8; RU: wt n = 5, DstacΔSH3.1 n = 5; RV: wt n = 9, DstacΔSH3.1 n = 9; each dot represents one cDNA gel band).