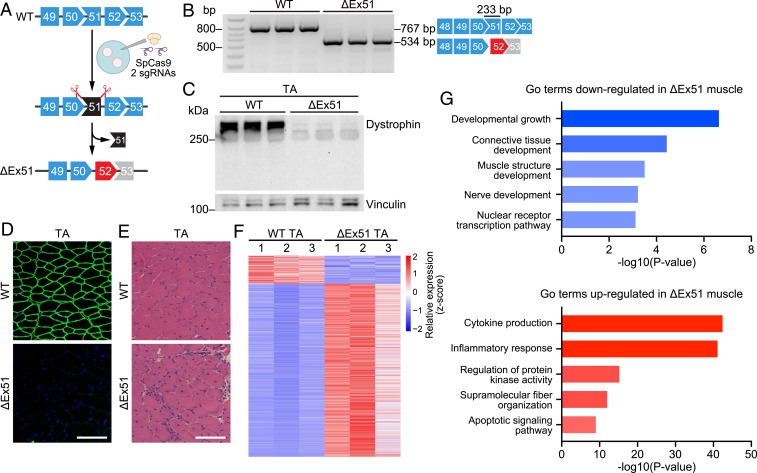
Fig. 1.
Creation and analysis of ΔEx51 Dmd mice. (A) CRISPR-Cas9 editing strategy used for generation of mice with dystrophin exon 51 deletion (ΔEx51). Mouse oocytes were injected with SpCas9 and 2 sgRNAs flanking exon 51. Upon deletion of exon 51, exon 52 (red) becomes out of frame with exon 50. (B) RT-PCR analysis of TA muscle to validate deletion of exon 51 (233-bp length). RT-PCR primers were in exons 48 and 53, and the amplicon size was 767 bp for WT mice and 534 bp for ΔEx51 mice. RT-PCR products are schematized on the right (n = 3). (C) Western blot analysis showing loss of dystrophin expression in the TA of ΔEx51 mice. Vinculin is the loading control (n = 3). (D) Dystrophin staining of the TA of WT and ΔEx51 mice. Dystrophin is shown in green. Nuclei are marked by DAPI stain in blue. (Scale bar, 100 μm.) (E) H&E staining of the TA of WT and ΔEx51 mice. Note extensive inflammatory infiltrate and centralized myonuclei in ΔEx51 muscle. (Scale bar, 100 μm.) (F) Heat map showing z-score–transformed expression of the differentially expressed genes between WT and ΔEx51 TA muscle (n = 3). (G) Selected top GO terms enriched in down- and up-regulated genes in ΔEx51 TA muscle.