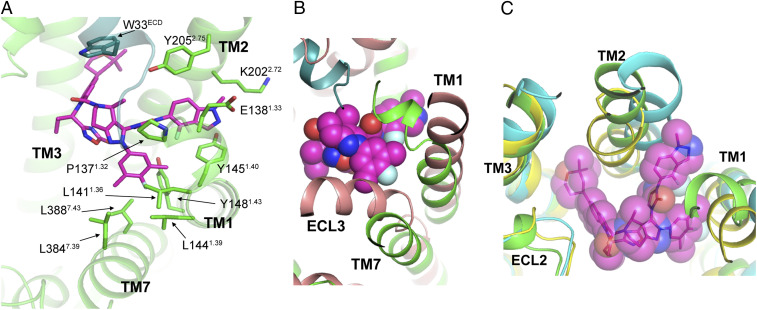
Fig. 3.
LY3502970 binding and the conformation of TM1, TM2, and TM7 of the GLP-1R. (A) LY3502970 (magenta) interactions with TM1, TM2, and TM7 of GLP-1R (green). Residues and ligands are shown by sticks. (B) TM1, TM7, and ECL3 of the inactive state structure of GLP-1R (salmon, PDB ID code 6LN2) need to shift to interact with LY3502970 (magenta sphere). (C) Unique conformation of TM1 and TM2 to accommodate LY3502970. The structures of GLP-1R bound to LY3502970, GLP-1 (yellow, PDB ID code 6VCB), and TT-OAD2 (cyan, PDB ID code 6ORV) are aligned. Other peptide bound GLP-1R structures (GLP-1, PDB ID code 5VAI; ExP5, PDB ID code 6B3J; peptide 5, PDB ID code 5NX2) are not shown here, but their TM1, TM2, and TM3 conformations are very similar to GLP-1 bound structure 6VCB in yellow.