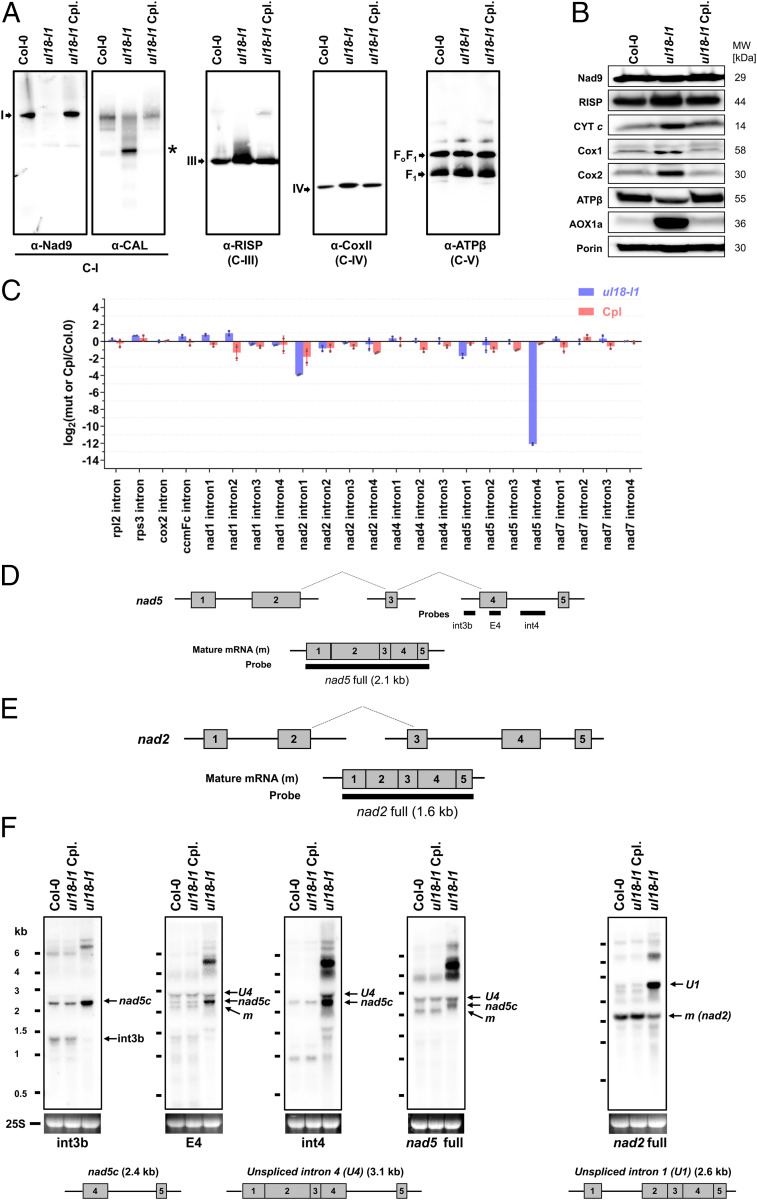
Fig. 4.
ul18-l1 plants are defective in complex I and are impaired in nad5 intron 4 and nad2 intron 1 splicing. (A) Immunodetection of respiratory complexes in blue native polyacrylamide gel electrophoresis blots made from mitochondrial extracts of WT (Col-0), ul18-l1 mutant, and complemented (Cpl) plants. Antibodies to Nad9 and CAL proteins were used to detect fully assembled and assembly intermediates of complex I (C-I), respectively. Complex III (C-III) was detected with antibodies to the RISP protein, complex IV (C-IV) with anti-Cox2 antibodies, and complex V (C-V) with anti-ATPβ antibodies (see SI Appendix, Table S2 for details). Respiratory complexes are designated by Roman numerals, and the asterisk designates a complex I assembly intermediate. (B) SDS/PAGE immunoblots performed on total mitochondrial protein extracts prepared from the indicated genotypes (see A for details) and probed with antibodies to subunits of respiratory complex I (Nad9), complex III (RISP), complex IV (Cox1 and Cox2), the complex V (ATPβ), cytochrome c (CYT c), and the alterative oxidase (AOX). Porin was used as protein loading control. (C) qRT-PCR analysis measuring the splicing efficiencies of mitochondrial introns in ul18-l1 (blue bars) and complemented (Cpl, red bars) plants. The bars depict log2 ratios of splicing efficiencies in ul18-l1 and complemented plants to the wild type (Col-0). Three technical replicates and two independent biological repeats were used for each genotype. SD are indicated. (D) Schematic representation of the three nad5 precursor transcripts, which are fused by two trans-splicing events to produce the mature (m) nad5 mRNA. Gray boxes indicate exons. Introns and 5′ and 3′ UTRs are shown as thin lines. The probes used to interpret RNA gel blot results are also indicated. (E) Schematic representation of nad2 precursors and mature mRNA, using the same iconography as in D. (F) RNA gel blots showing the accumulation profiles of nad5 and nad2 transcripts in the ul18-l1 mutant compared to WT (Col-0) and complemented plants (Cpl). Used probes are indicated below hybridization results. Ethidium bromide staining of the 25S ribosomal RNA serves as a loading control. Schematic representations and sizes of the nad5 transcript with unspliced intron 4 (nad5 U4), nad5c, and nad2 transcript with unspliced intron 1 (nad2 U1) are depicted below the blots to help interpreting the results. The apparent size of nad5 U4 is smaller than expected due to a congestion by near-sized 25S rRNA (24). RNA marker sizes are indicated (kilobases). E, exon; int, intron; m, mature mRNAs.