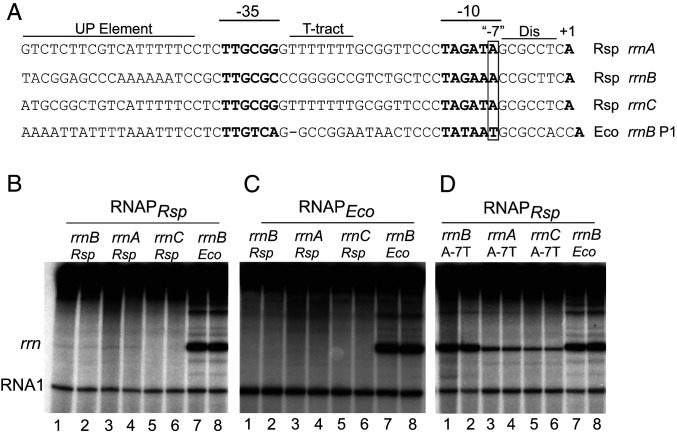
Fig. 1.
Activity of R. sphaeroides rRNA promoters with either R. sphaeroides RNAP or E. coli RNAP. (A) Sequences of the three R. sphaeroides rRNA promoters, rrnA, rrnB, and rrnC, from −57 to the transcription start site, +1, and the E. coli rrnB P1 promoter. The discriminator (Dis) region, T-tract sequence, and UP element are indicated, in addition to the −10 and −35 RNAP recognition hexamers and the transcription start site, which are in bold. The last bp in the −10 element is referred to as the “−7” position (boxed), although it is 8 or 9 bp rather than 7 bp upstream from the transcription start site due to the larger-than-consensus number of bp between the −10 element and the TSS in these promoters. (B) In vitro transcription of the indicated rRNA promoters from plasmid templates with R. sphaeroides RNAP in buffer containing 170 mM NaCl (SI Appendix, Expanded Materials and Methods). Duplicate lanes are shown for each promoter. The RNA I promoter and transcript are part of the plasmid replication control system (SI Appendix, Expanded Materials and Methods). (C) In vitro transcription of the indicated promoters as in B, but with E. coli RNAP. (D) In vitro transcription with R. sphaeroides RNAP as in B, but with the A-7T promoter variants of the three R. sphaeroides rRNA promoters or with the wild type E. coli rrnB P1 promoter. A higher concentration of R. sphaeroides RNAP (50 nM) than E. coli RNAP (10 nM) was used to ensure that the absence of transcription from the R. sphaeroides rRNA promoters was not a result of limiting RNAPRsp. Robust, approximately equivalent transcription was observed from the E. coli rrnB promoter at 50 nM RNAPRsp and 10 nM RNAPEco (B–D).