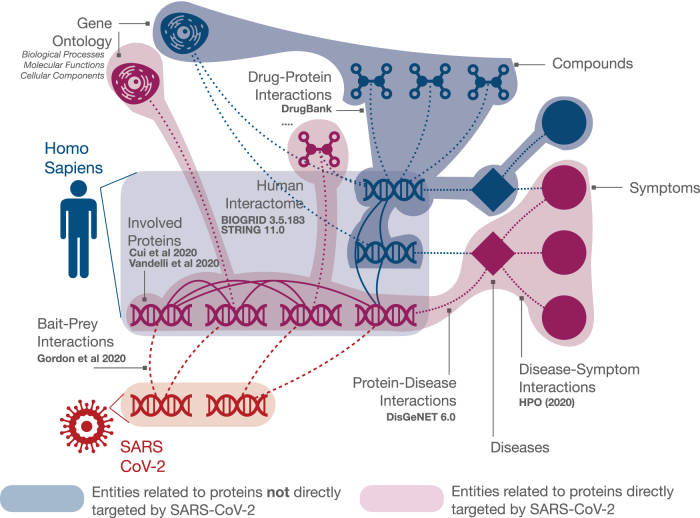
FIG. 1.
Linking genotype to phenotype in SARS-CoV-2–Homo sapiens molecular interactions. We build a highly reliable map of the human interactome and focus on the subset of human proteins that were shown to putatively interact with the virus in the literature, both through experimental protein interaction assays,7 through structure-based predictions,9 and based on similarity of the proteins to other coronaviruses proteins.8 The COVID-19 PPI network is enriched by biological information related to each involved protein (GO terms), as well as by an extensive data set of drug–protein interactions obtained by integrating different repositories. Finally, the system is enriched with phenotype information about diseases and symptoms, allowing us to include disease–symptom and protein–disease associations. Different icons represent different entities: genes, diseases, compounds, and symptoms are represented by DNA fragments, diamonds, chemical structures, and circles, respectively. Purple shaded area and purple icons represent entities associated with genes of human proteins directly targeted by SARS-CoV-2, whereas blue shaded area and blue icons denote entities related to genes of human proteins indirectly targeted by SARS-CoV-2 through human PPI. Cell icons represent GO terms, including biological processes, molecular functions, and cellular components. Solid lines highlight human PPIs and dotted lines represent other types of interactions between different entity types. See the text for details. GO, Gene Ontology; PPI, protein–protein interaction.