Fig. 2.
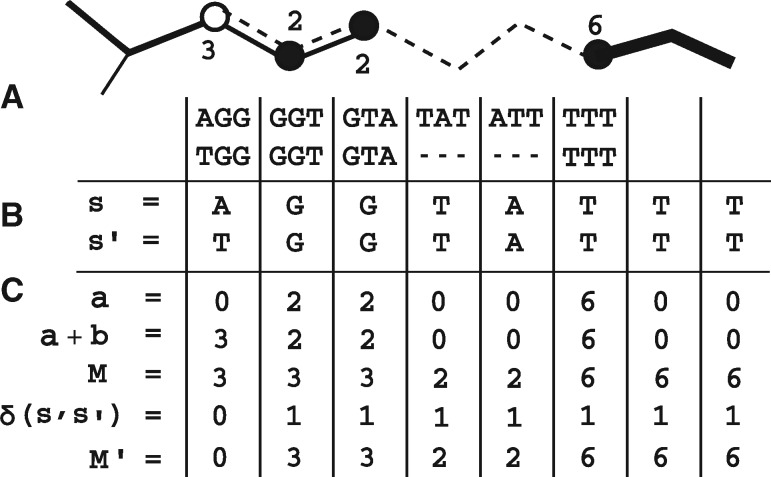
Scoring procedure for an example graph mapping. An example mapping of a reference s (dashed line) to a graph (solid lines), with exact matches (black circles) and inexact matches (white circles), is shown (A), with string representation of the path, s′ (B). Firstly, a weight array a is retrieved for which ai gives the weight of the ith reference k-mer in the wDBG, where gaps are given a weight of zero. These exact matches are then extended with bridges b to inexact matches. Next, per-base weights M are calculated such that each base is given the greatest weight of any k-mer that includes it, which also functions to assign weights to terminal characters of a string (or substring before a gap) that do not have k-mers (such as ‘TA’ at the fourth position). Finally, the array M′ is computed as , where if , and zero otherwise. For our classifier, we chose to multiply the sum of this array by the fraction of non-gap (non-zero) positions, in order to penalize high weight, high gap mappings