Figure 3. The Diverse Transcription Factor (TF) Repertoire of Mature Salivary Glands May Shape Hotspots of Salivary-Gland-Specific Expression across the Genome.
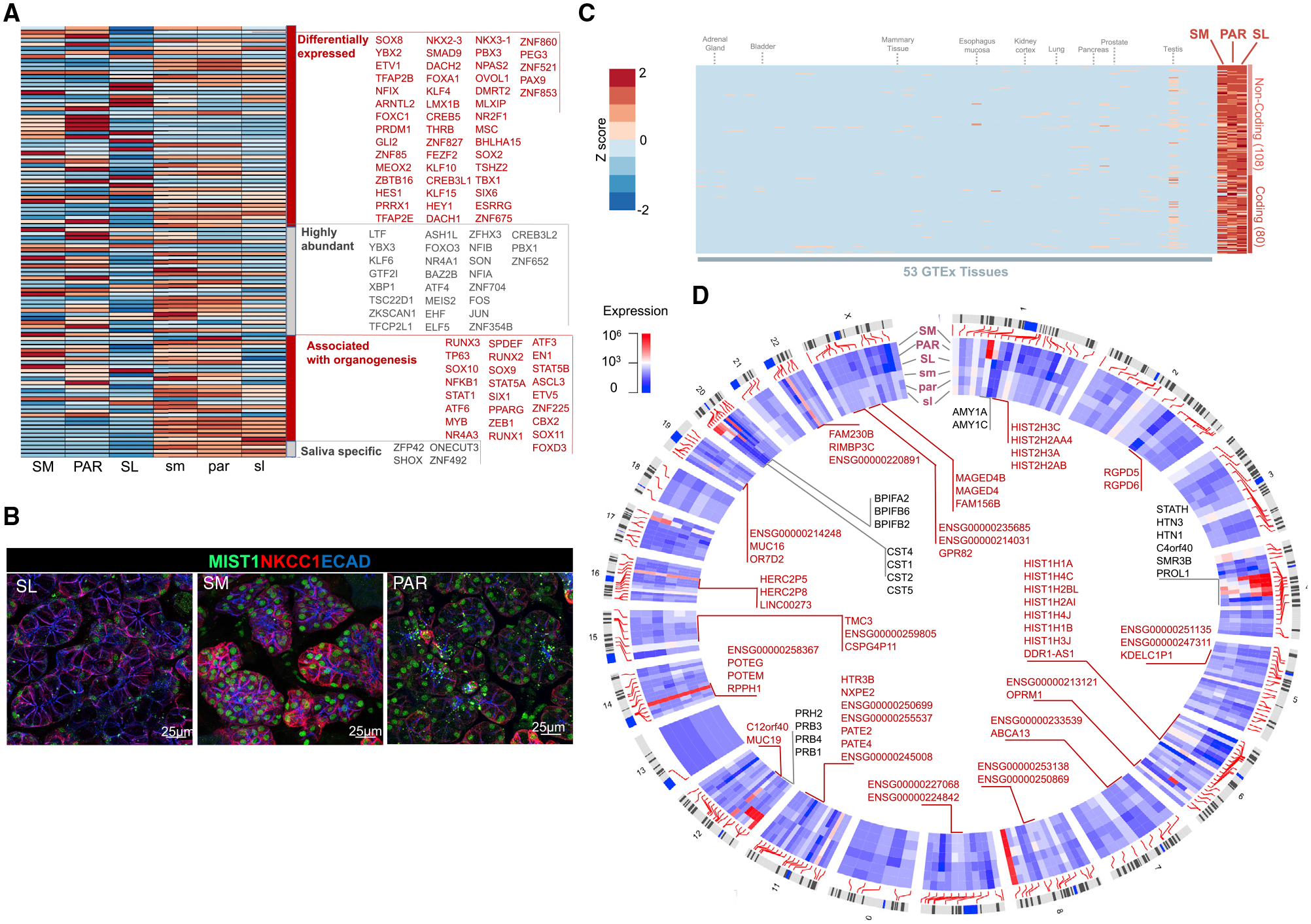
(A) Heatmap of expression levels of TF genes (as listed in TF2DNA database; Pujato et al., 2014) across fetal and mature salivary gland tissues. Four categories of TFs are shown in the heatmap: TFs that are (1) differentially expressed (p < 0.0001) among mature glands, (2) abundant (>2,000 NCs) in adult or fetal glands, (3) previously associated with organogenesis, and (4) salivary gland specific (that is, >100 NCs in the salivary glands but negligible expression in all 53 GTEx tissue [<10 transcripts per million (TPM)]). LTF, a secreted protein in saliva, is listed here because one of its isoforms, delta lactoferrin, displays TF activity (He and Furmanski, 1995; Mariller et al., 2007).
(B) Immunofluorescent analysis of TF BHLHA15/MIST1 in adult glandular tissues. The SM and PAR cells are highly enriched for MIST1 compared with the SL cells. NKCC1/SLC12A2, Na-K-Cl cotransporter 1. Scale bar, 25 μm.
(C) Heatmap of expression levels of genes in mature salivary glands (>100 NCs) that show negligible expression in other tissues and organs (expression in all 53 GTEx tissues < 10 TPM). The specific tissues in the GTEx database used for this analysis are listed in Table S5. Epithelial or secretory tissues and organs important for comparison to salivary glands are indicated on top of the heatmap. Deviation from the mean expression for each column is shown as a Z score with a scale similar to that used in (A).
(D) Circos plot showing the locations of genes with salivary-gland-specific expression. These genes show considerable expression in the salivary glands (>100 NCs) but negligible expression in all 53 GTeX tissues. Clusters of genes located within 1 Mb of one another are pointed out with gene names inside the Circos plot. Genes that previously had not been reported within the context of salivary glands are indicated in red.
