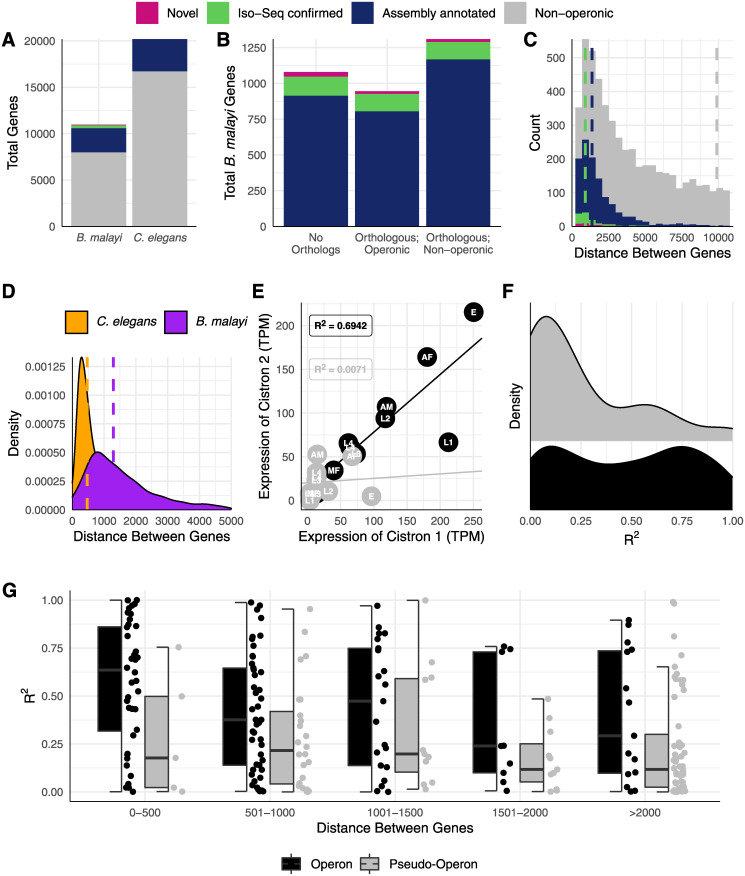
Fig 5. Identification and validation of over a hundred operons in B. malayi.
(A) Iso-Seq identified new operons and validated assembly-annotated operons in B. malayi, which has a similar proportion of its genome arranged in operons as C. elegans. (B) Most B. malayi genes in operons have orthologs in C. elegans that are not in operons (39%), while 32% do not have orthologs, and 28% have orthologs that are also in operons. (C) Intercistronic distances between genes in operons are much shorter than intergenic distances between non-operonic genes (note that green and magenta median lines overlap). Operons identified via Iso-Seq (a median of 915.0 nucleotides for new operons, 906.5 for validated operons) on average had shorter intercistronic distances than those already annotated in the Bmal-4.0 assembly (1362.0). (D) B. malayi operons on average have larger (median = 1289 contrasted to 467 for C. elegans) and a broader range (interquartile range of 1422.5 contrasted to 700.5 for C. elegans) intercistronic distances than those in C. elegans. (E) An example of linear fits to staged RNA-seq data from genes in a pseudo-operon (non-operonic neighboring genes) and an operon. (F) Linear fits better explain the relationship between expression of neighboring genes in operons than neighboring genes in pseudo-operons, and (G) this contrast does not stratify by intergenic distance.