Figure 1.
Integration of Transcriptional Inputs over Time as a Mechanism for Cell Diversification
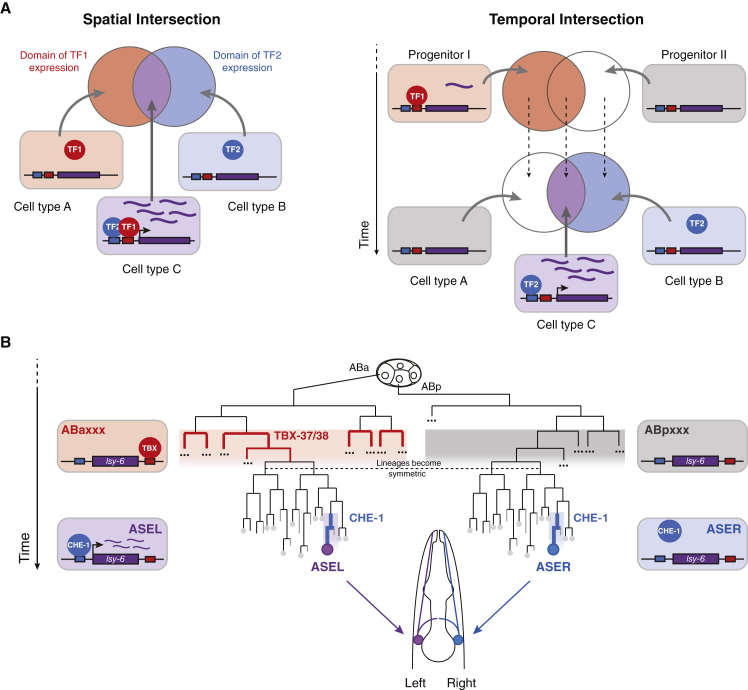
(A) Left. Schematic of the classic mode of combinatorial TF action based on spatial intersection of two TFs. Circles denote the partially overlapping domains of expression of two TFs. Boxes represent cells within those domains. Expression of TF1 or TF2 alone is not sufficient to activate a gene relevant for cell identity (in purple) but action of both TFs together is. Right. Schematic of the concept of temporal intersection in which two TFs are required for transcriptional activation, but separated in time. TF1 is expressed transiently in a subset of progenitor cells, while TF2 is expressed in a subset of cells undergoing terminal differentiation. Only a cell expressing TF2 that derived from a progenitor that expressed TF1 will activate the purple gene. Therefore, the outcome of the presence of TF2 in a cell depends on the transcriptional history of that cell, and this mechanism can contribute to cellular diversification.
(B) Schematic of the lineage histories of the left and right ASE neurons (ASEL and ASER), which derive from the ABa and ABp blastomeres, respectively, at the 4-cell stage of embryogenesis. Both neurons express the terminal TF CHE-1 at the endpoint. CHE-1 is necessary for lsy-6 expression, but only ASEL expresses lsy-6. Asymmetric expression of lsy-6 is genetically dependent on the transiently expressed TBX-37/38, which have a predicted binding site in the lsy-6 locus.