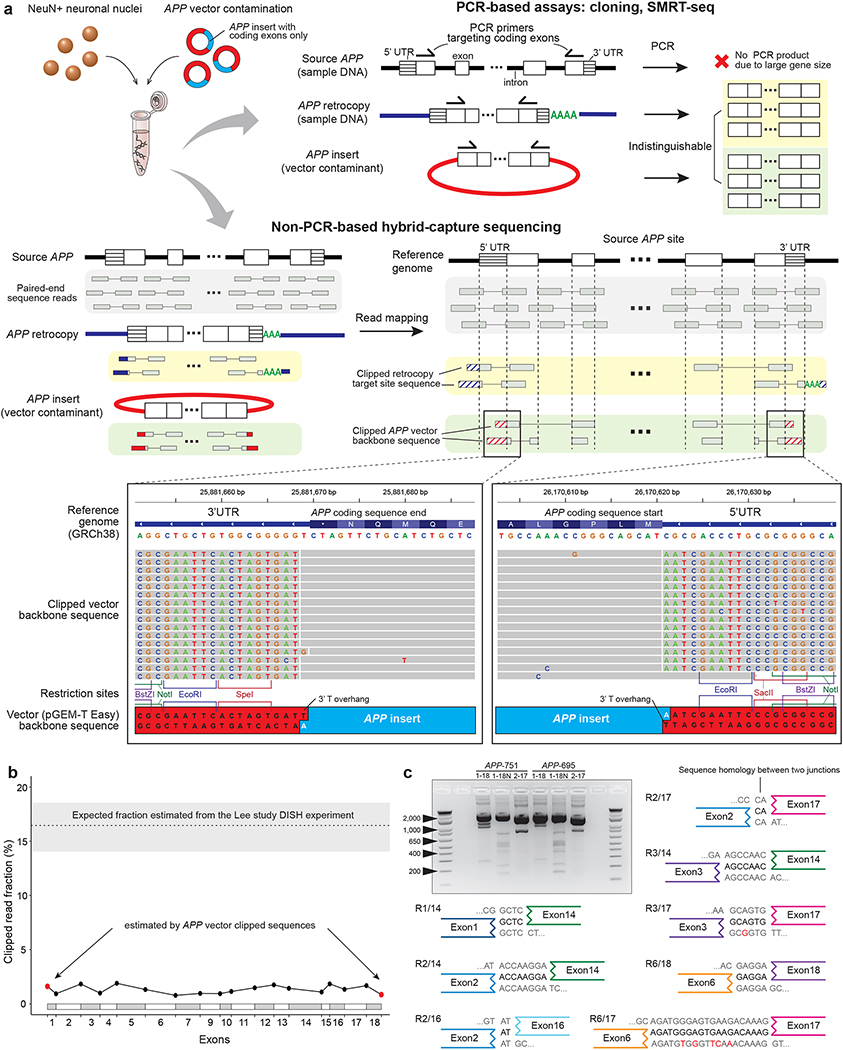
Figure 1. APP vector contamination in the Lee study.
a. APP vector contamination and its manifestation in genome sequences. PCR-based assays in the Lee study fail to distinguish between APP retrocopy and vector APP insert. Hybrid-capture sequences from the Lee study shows clipped reads with a vector backbone sequence (pGEM-T Easy), including restriction sites at the multiple cloning site and a 3’ T-overhang. b. Estimated fractions of cells with APP gencDNA at the exon junctions in the Lee hybrid-capture data. All exon junction fractions (black dots) are comparable to the fraction at the coding sequence ends with vector backbone sequences (red dots). The dotted line on the top represents the conservative estimate of expected fraction based on the Lee DISH experiment (see Supplementary Methods); shaded area, 95% confidence interval. c. Electrophoresis and sequencing of PCR products from the vector APP inserts (APP-751/695) showing novel APP variants as artifacts. Eight out of 12 IEJs found both in our APP vector PCR sequencing and the Lee study RT-PCR results are shown (see also Extended Data Fig. 3).