Fig. 2.
The Caenorhabditis elegans natural diversity resource (CeNDR) provides insights into genetic and phenotypic variation of natural C. elegans population.
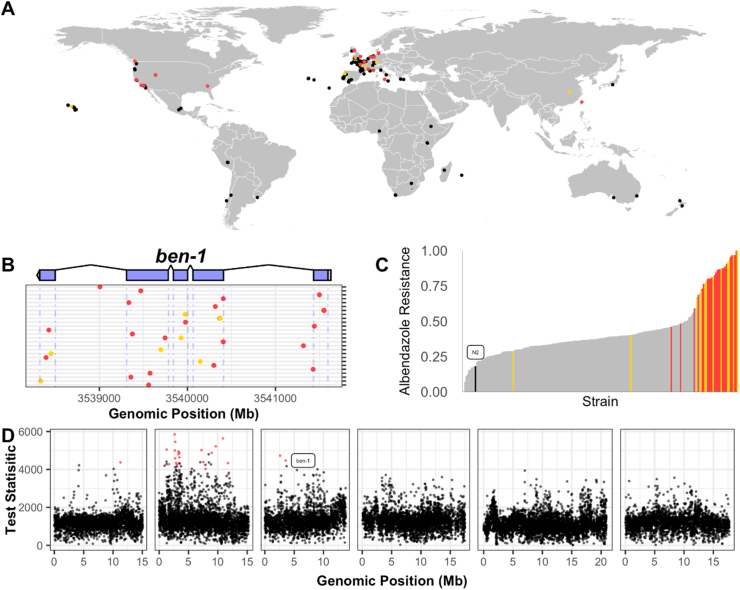
(A) The map shows global origin of C. elegans strains that are available to the community through CeNDR. Overall, 249 genetically diverse strains were collected over the past 50 years by researchers and citizen-scientists from different regions and substrates all over the world. All isolated strains were cryopreserved and genome sequenced. (B) Genomic variation in anthelmintic drug targets can be queried with the CeNDR Variant Browser tool (https://www.elegansvariation.org/data/browser/). As an example, the genetic variation in the beta-tubulin gene ben-1, a major resistance gene for benzimidazoles, is shown. Genetic variants with predicted moderate (e.g. missense variants) or high effects (e.g. frame shift variants) are displayed in yellow or red, respectively. (C) CeNDR strains vary in their phenotypic response to anthelmintic drugs. As an example, the relative resistance to albendazole (ABZ) is shown (Hahnel et al., 2018). Each bar represents a single CeNDR strain, included in an ABZ exposure assay, sorted by their relative ABZ resistance. Strains that have ben-1 variants with predicted moderate or high effects are colored in yellow and red, respectively. Strains similar to the N2 reference genome with respect to the ben-1 locus are shown in grey. Distribution of strains with a ben-1 variant indicate a correlation between ben-1 and ABZ resistance in C. elegans wild isolates. (D) Phenotype and genotype data of wild isolates can be used to identify genomic regions or genes that correlate with an observed resistance phenotype. The example shows the Manhattan plot of a genome-wide association study performed to identify genetic determinants of BZ resistance in CeNDR strains (Hahnel et al., 2018). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)