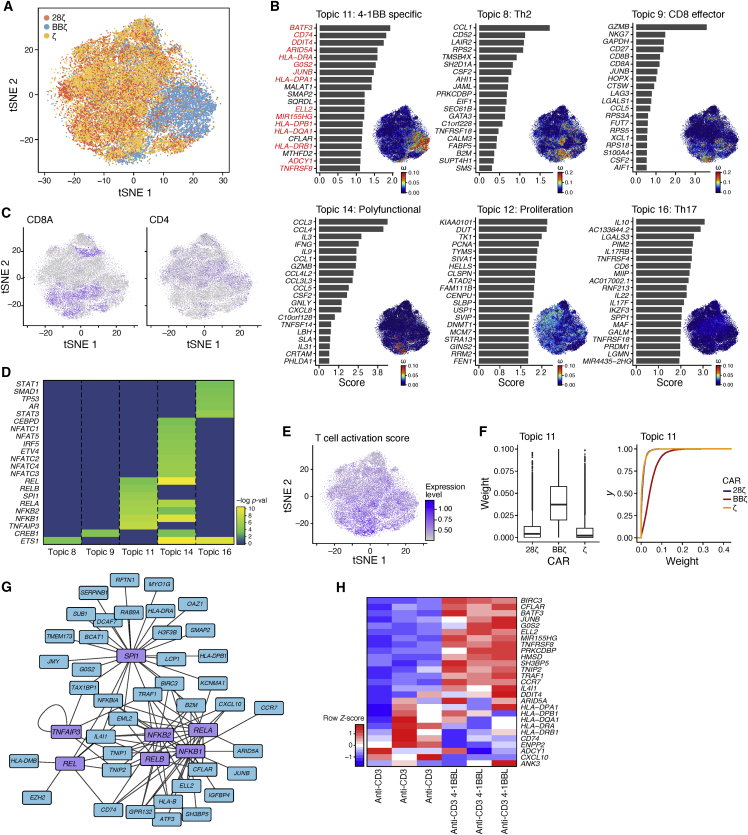
Figure 5.
Antigen-Specific Activation of 4-1BB CAR T Cells Induces a Distanced Program with Additional Genes Networks Than 4-1BB Ligand-Mediated Triggering of 4-1BB
Single-cell expression profiles of CAR T cells after 24 h of stimulation with Nalm6 cells were normalized and aligned across two donors. (A) tSNE plot of single-cell expression profiles (dots) colored by CAR T cell construct. (B) Results of latent Dirichlet allocation on T cells with 16 topics and a tolerance parameter of 0.1 (see Materials and Methods). For each topic shown, there is a bar plot of top-scoring genes (y axis), ranked by a uniqueness score. The genes in topic 11, which were also found as DE and upregulated (adj-p < 0.05) in BBζ versus 28ζ CAR T cells at any time point from bulk RNA-seq data, are in red. Each cell (dot) of the tSNE is colored by the weight of the topic (see also Figure S8 and Table S7). (C) Degree of CD4 and CD8A expression in cells plotted in the tSNE as in (A). (D) Gene regulators discovered using network analysis of the top 100 genes in each topic. Transcription factors with a statistical association (FDR < 0.1) were plotted by −log(pval) for each topic of interest (see also Table S8). (E) T cell activation signature expression level in cells plotted in the tSNE as in (A) (see Table S3 for activation signature). (F) Box-and-whisker plot and cumulative distribution plot of the topic 11 weights per cell across the different CAR T cell groups. (G) Network of predicted transcription factors identified and the genes they regulate in the 4-1BB program (topic 11). (H) UT T cells were stimulated for 24 h with irradiated K562 expressing αCD3 with or without 4-1BBL at a 1:1 effector-to-target cell ratio. Heatmap of 4-1BB CAR signature genes in UT T cells stimulated with and without 4-1BBL (normalized by row) gene signatures. N = 3 normal donors.