Extended Data Figure 6. Transposable elements in SIP formation.
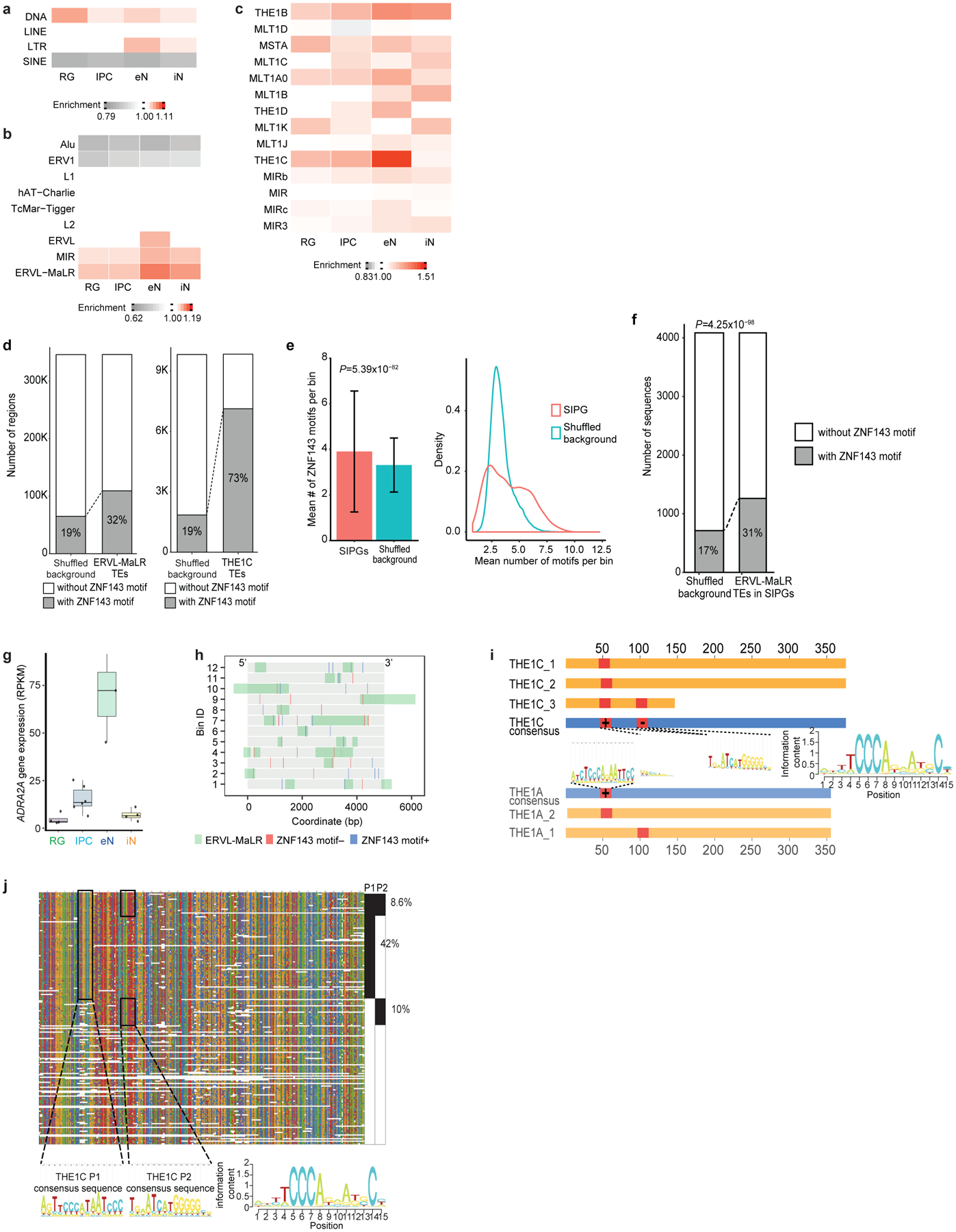
(a-c) Enrichment of TEs at the class (a), family (b), and subfamily (c) levels in SIPGs for each cell type. Only TE families occupying more than 1% of the genome are shown in (b). Only TE subfamilies from the MIR and ERVL-MaLR TE families occupying more than 0.1% of the genome are shown in (c). (d) Both ERVL-MaLR TEs (left, 32% versus 19% of sequences, P < 2.2*10−16, binomial test, two-tailed) and THE1C TEs (right, 73% versus 19% of sequences, P < 2.2*10−16, binomial test, two-tailed) are enriched over background sequences for ZNF143 motifs in eNs. (e) ZNF143 motifs are enriched at SIPGs in eNs (left, P = 5.39×10−82, two-sample t-test, two-tailed, n = 8,894 distal interacting regions). Means are indicated and error bars represent the SEM. Distributions comparing the number of ZNF143 motifs per bin for actual versus shuffled SIPGs are shown (right, P < 2.2*10−16, Kolmogorov-Smirnov test, two-tailed, n = 638 SIPGs). (f) ERVL-MaLR TEs in SIPGs are enriched over background sequences for ZNF143 motifs in eNs (31% versus 17% of sequences, P = 4.3×10−98, binomial test, two-tailed). (g) Box plots showing elevated ADRA2A gene expression in eNs. The median, upper and lower quartiles, minimum, and maximum are indicated. (h) Illustration of the 12 distal interacting regions containing ERVL-MaLR TE-localized ZNF143 motifs in the ADRA2A SIPG. ZNF143 motifs are colored by strand. The bin numbers correspond to Fig. 3j. (i) Conservation of ERVL-MaLR TEs in the ADRA2A SIPG. Blue bars indicate consensus sequences, yellow bars indicate ERVL-MaLR TEs, and red bars indicate ZNF143 motifs. (j) Alignment of THE1C TEs in the human genome to their consensus sequence. The THE1C subfamily contains two ZNF143 motifs, one at positions 47–61 (P1), and another at positions 96–110 (P2).
