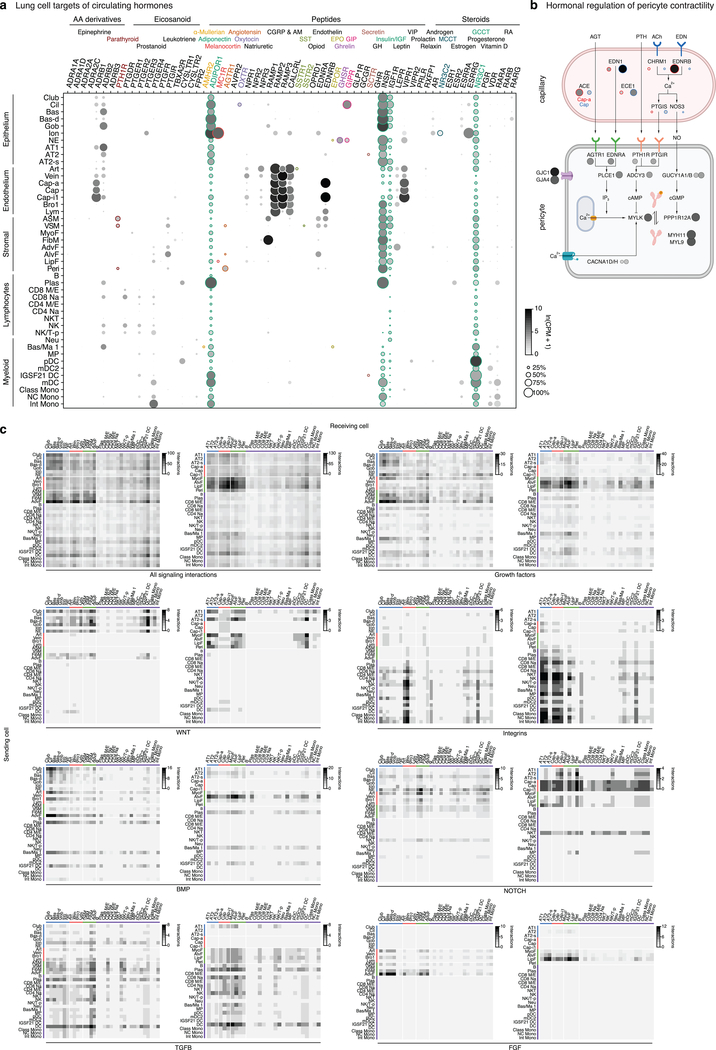
Extended Data Figure 6. Lung cell targets of circulating hormones and local signals.
a, Dot plot of hormone receptor gene expression in lung cells (SS2 dataset). Type and name of cognate hormones for each receptor are shown at top. Teal, broadly-expressed receptors in lung; other colors, selectively-expressed receptors (<3 lung cell types). Small colored dots next to cell type names show selectively targeted cell types. AA, amino acid; CGRP, Calcitonin gene-related peptide; AM, adrenomedullin; SST, somatostatin; EPO, erythropoietin; GIP, gastric inhibitory peptide; GH, growth hormone; IGF, insulin-like growth factor; MCCT, mineralocorticoid; GCCT, glucocorticoid; RA, retinoic acid. b, Schematic of inferred pericyte cell contractility pathway and its regulation by circulating hormones (AGT, PTH) and capillary-expressed signals (EDN, NO). Dots show expression of indicated pathway genes: values at left (outlined red) in each pair of dots in capillary diagram (top) show expression in Cap-a cells (aerocytes) and at right (outlined blue) show expression in general Cap cells (SS2 dataset). Note most signal genes are preferentially expressed in Cap relative to Cap-a cells. c, Heatmaps showing number of interactions predicted by CellPhoneDB software between human lung cell types located in proximal lung regions (left panel in each pair) and distal regions (right panel) based on expression patterns of ligand genes (“Sending cell”) and their cognate receptor genes (“Receiving cell”) (SS2 dataset). The pair of heatmaps at upper left show values for all predicted signaling interactions (“All interactions”), and other pairs show values for the indicated types of signals (growth factors, cytokines, integrins, WNT, Notch, Bmp, FGF, and TFGB). Predicted interactions between cell types range from 0 (lymphocyte signaling to neutrophils) to 136 (AdvF signaling to Cap-i1). Note expected relationships, such as immune cells expressing integrins to interact with endothelial cells and having higher levels of cytokine signaling relative to their global signaling, and unexpected relationships, such as fibroblasts expressing majority of growth factors and lack of Notch signaling originating from immune cells. For more details on statistics and reproducibility, please see Methods.