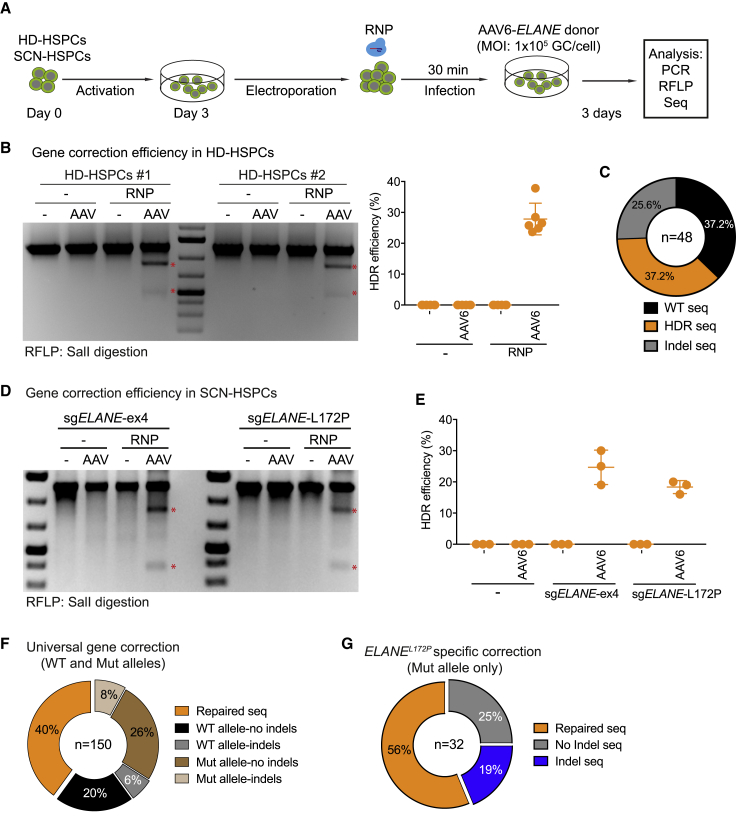
Figure 2.
Efficiency of ELANE Correction in HD- and SCN Patient-Derived CD34+ HSPCs
(A) Schematic of experiment to assess the efficiency of the ELANE correction in either HD-HSPCs or SCN-HSPCs. (B) Left: SalI-RFLP assays showing the HDR events using universal exon 4-based ELANE correction (sgELANE-ex4) in HD-HSPCs from two individuals (HD#1 and HD#2). Stars indicate SalI-cleaved fragments. Right: quantification of three independent duplicate experiments. Data are represented as mean ± SD. (C) Pie chart showing WT, HDR, and indel events of the targeted ELANE locus in HD-HSPCs. (D) SalI-RFLP assays showing efficiency of universal exon 4-based ELANE- (left) or ELANEL172P-specific (right) correction in SCN-HSPCs. Scars indicate SalI-cleaved bands. (E) Quantification of the correction efficiency of universal (sgELANE-ex4) and specific (sgELANE-L172P) systems based on three independent experiments. Data are represented as mean ± SD. (F) Pie chart showing repaired, WT, and mutant (Mut) alleles with or without indel events of the targeted ELANE locus in SCN-HSPCs undergoing universal exon 4-based ELANE gene correction. (G) Pie chart showing repaired and Mut allele with or without indel events of the targeted ELANE locus in SCN-HSPCs undergoing the ELANEL172P-specific correction. See also Figures S3–S5.