Figure 4.
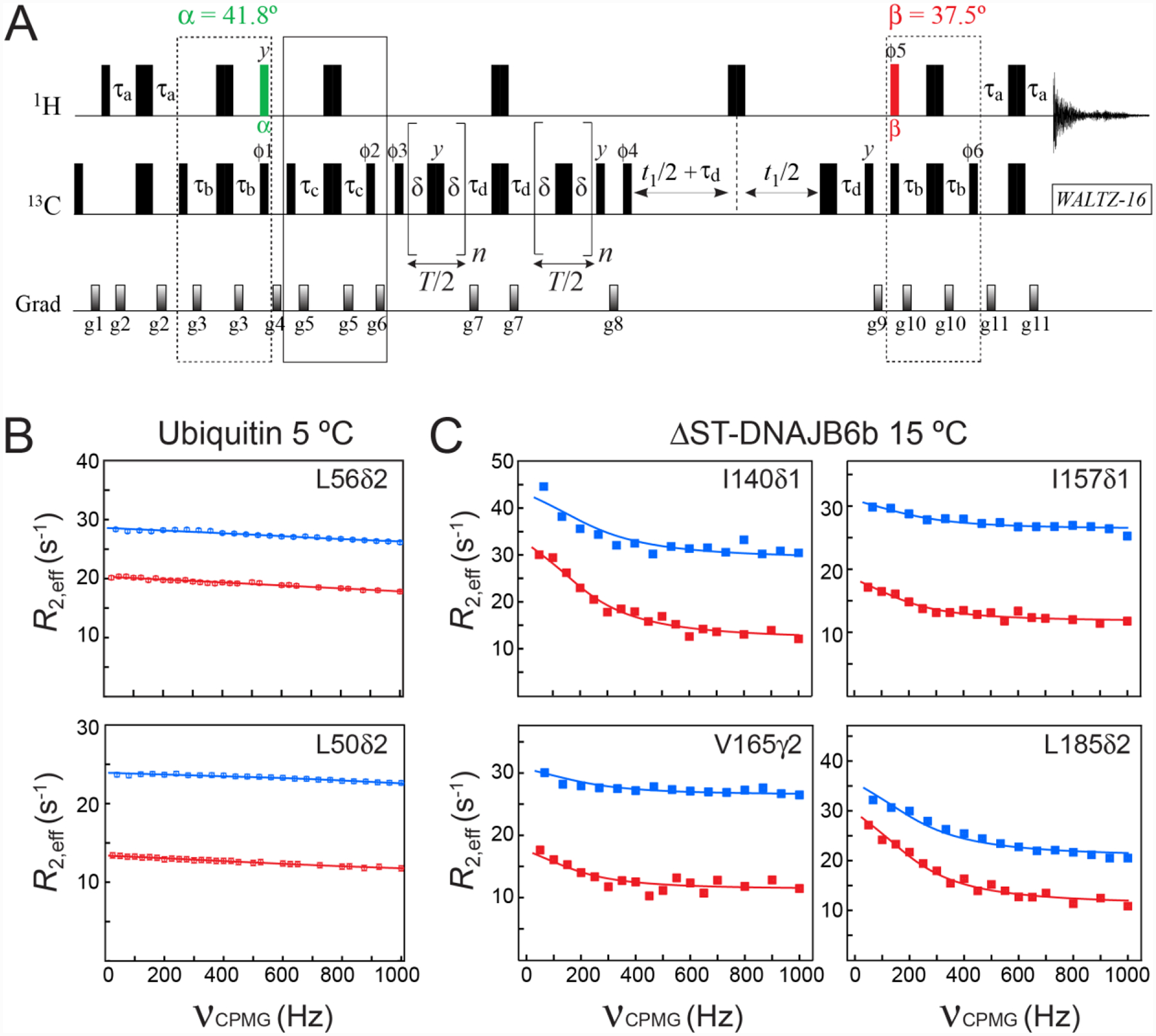
(A) Pulse scheme for methyl 13C SQ CPMG relaxation dispersion experiment with optimal selection of slow-relaxing 13C transitions. All the parameters of the scheme are as described in Figures 2A and 3A. The delay τd = 1/(4JHC) = 2.0 ms. T is the CPMG constant time relaxation delay. Even number of CPMG cycles (180° 13C pulses) n should be used for each relaxation period T/2. The durations and strengths of pulsed-field gradients in units of (ms; G/cm) are: g1 = (1.0; 25), g2 = (0.5; 15), g3 = (0.3; 20), g4 = (1.5; 25), g5 = (0.25; 15), g6 = (1.2; 20), g7 = (0.5; 20), g8 = (0.6; 20), g9 = (0.8; −25), g10 = (0.3; 20), g11 = (0.5; 20). The phase cycle is: ϕ1 = 8(x),8(−x); ϕ2 = 4(x),4(−x); ϕ3 = 2(x),2(−x); ϕ4 = x,-x; ϕ5 = 8(x),8(−x); ϕ6 = 8(x),8(−x); receiver = (x, −x, −x,x, 2(−x,x,x, −x), x, −x, −x,x). Quadrature detection in t1 is achieved via States-TPPI incrementation of ϕ4. (B–C) Examples of methyl 13C relaxation dispersion profiles obtained for (B) ubiquitin at 5 °C, and (C) ΔST-DNAJB6b at 15 °C are shown in red. The corresponding profiles obtained with the scheme of Lundström et al. (2007) that does not select the slow-relaxing 13C transitions, are shown in blue. The best fits to a two-state exchange model are shown as continuous lines. In the case of ΔST-DNAJB6b, exchange occurs between the major monomeric state and a sparsely-populated, high-molecular-weight oligomer comprising 30–40 monomeric units (Karamanos et al. 2019). Sample conditions, the parameters of NMR experiments and the details of the fitting procedure are described in the ‘Materials and Methods’ section of the Supplementary Information.
