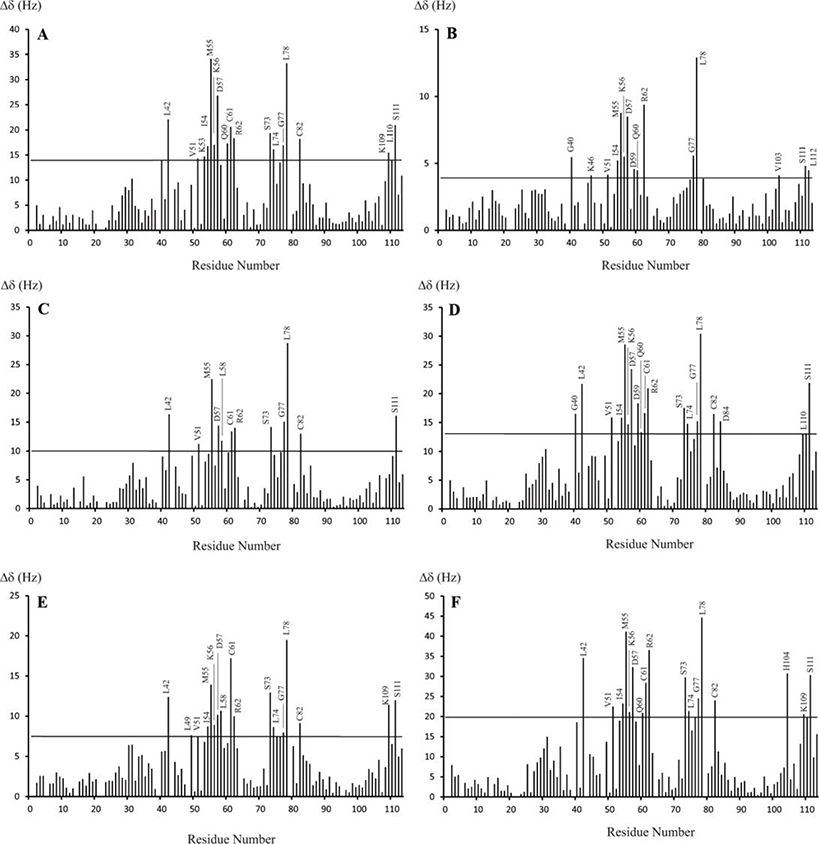
Figure 4. Chemical shift perturbation of 15N-S100A10-AnxA2Nter in complexes with CEACAM1-SF peptides.
The chemical shift changes between free 15N-S100A10-AnxA2Nter and in complex with six CEACAM1-SF peptides. The molar ratio between peptides and 15N-S100A10-AnxA2Nter for all complexes is 10:1 except for the complex with Ac-SW, which is 8.7:1. The thin line in each panel is the two times RMSD of the chemical shift changes between free 15N-S100A10-AnxA2Nter and 15N-S100A10-AnxA2Nter in complex with peptides. The peptide in complex is Ac-SW for A, Ac-SW-F/A for B, Ac-SW-T/E for C, Ac-SW-S/E for D, Ac-SW-TS/EE for E and Ac-LF for F. The sequences for each peptide are listed in Table 1. The CSP values in unit of HZ was calculated using the following equation. . The ΔωΝ and ΔωΗ are the nitrogen and proton chemical shift difference between free 15N-S100A10-AnxA2Nter and that in the mixture with molar ratio between peptides and 15N-S100A10-AnxA2Nter listed above.