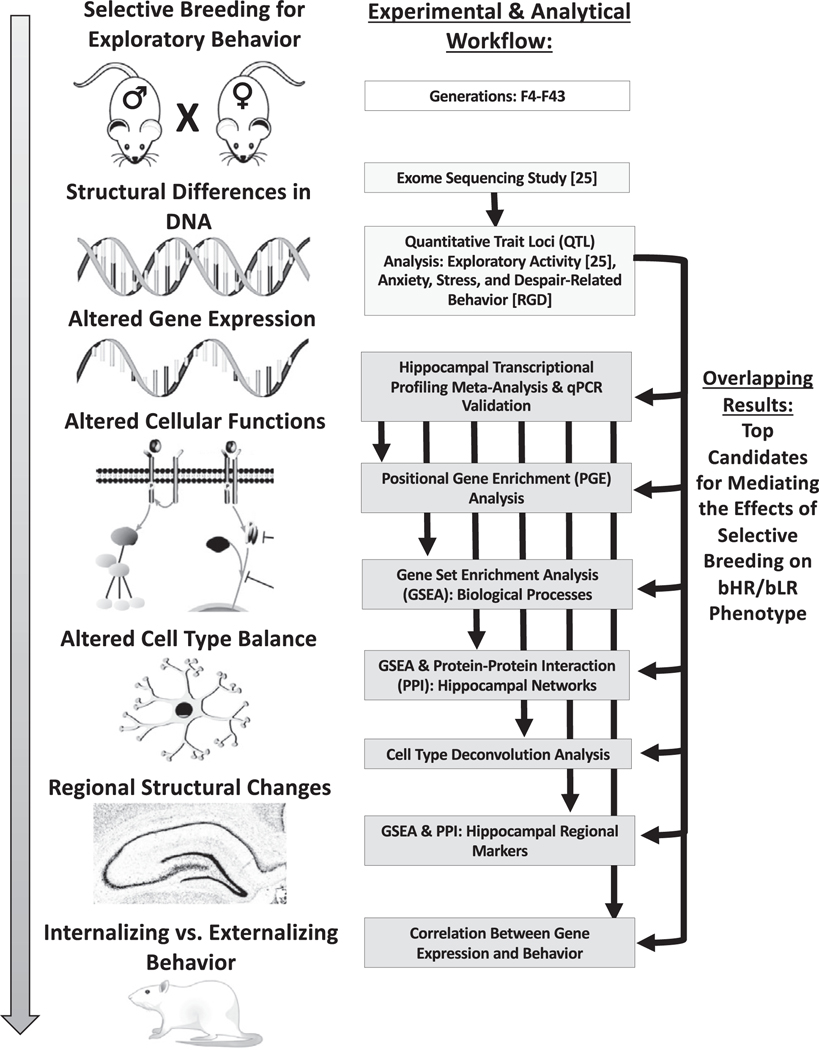
Figure 1.
An overview of the experimental and analytical workflow used to identify top candidate genes for mediating the effects of selective breeding on bHR/bLR phenotype. Left: Many generations of selective breeding based on exploratory locomotor behavior drove segregation of genetic variants that contribute to internalizing and externalizing behavior within our bHR and bLR rats. The effect of these variants on behavior is mediated by alterations in gene expression and cellular function, which produce local changes in cell type balance and structure within brain regions responsible for affective behavior such as the hippocampus. Right: Our concurrent genetic study used exome sequencing to identify genetic variants that segregated bHR/bLR rats in our colony and then used a sampling of those variants to locate regions of the genome (QTL) that might contribute to exploratory locomotor activity (25). Our current study used meta-analyses of hippocampal transcriptional profiling studies to identify bHR/bLR DE genes, pathways, cell types, and networks in development and adulthood. In our results, we emphasize DE genes that were 1) consistently DE across multiple developmental stages, 2) central to differential expression pathways, cell types, and networks, and 3) located near genetic variants that segregated bHR/bLR rats in our colony and/or within QTL for exploratory locomotion. These genes are the top candidates for mediating the effects of selective breeding on bHR/bLR phenotype. bHR, bred high-responder; bLR, bred low-responder; DE, differentially expressed; GSEA, gene set enrichment analysis; PPI, protein–protein interaction; qPCR quantitative polymerase chain reaction; QTL, quantitative trait loci.