Figure 4. Allele-specific methylation and accessibility in GM12878.
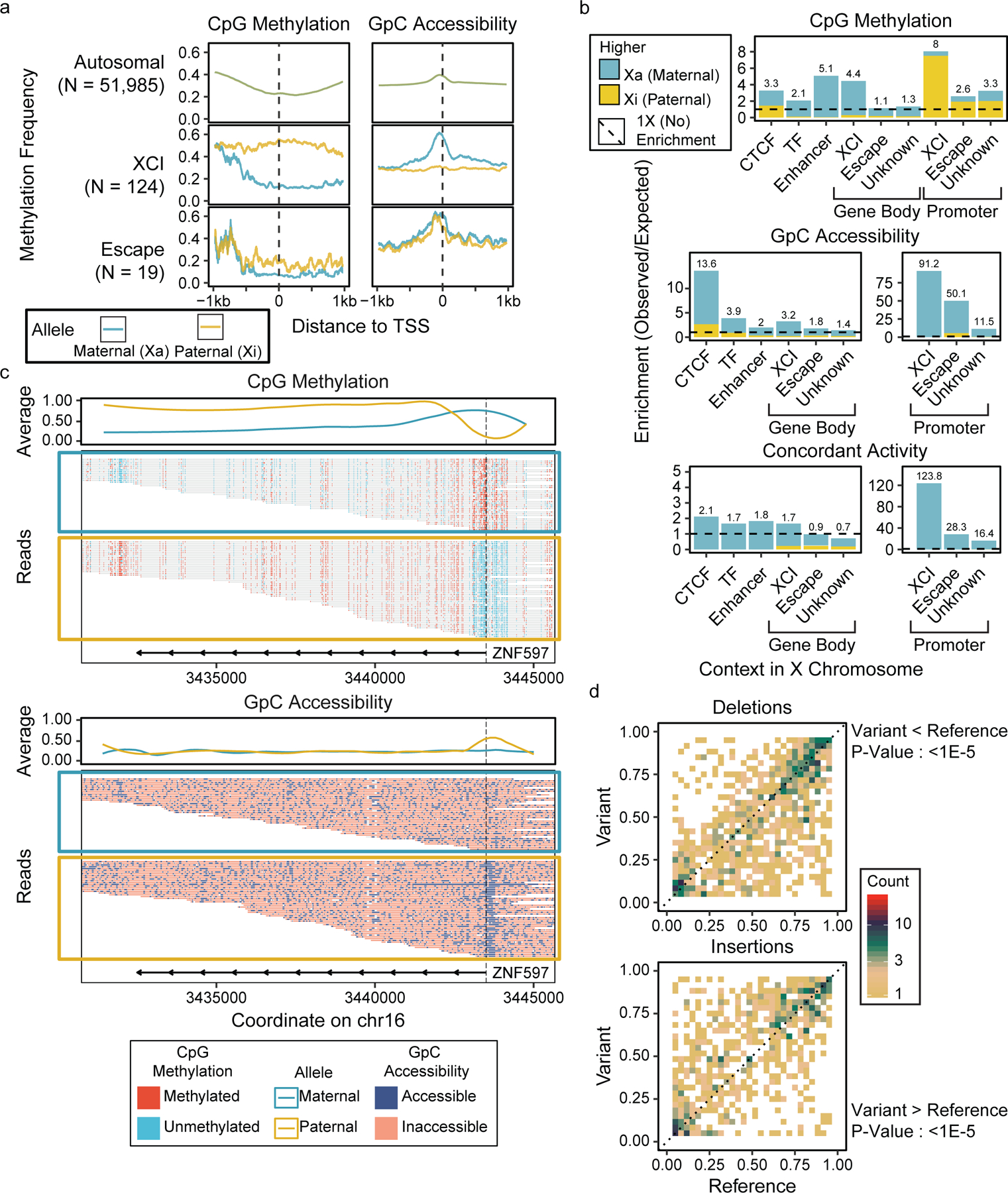
Methylation and accessibility were separated by parent of origin, and (a) metaplots of TSS methylation and accessibility were generated for each allele (b) Enrichments of DMRs and DARs were calculated at various genomic contexts, showing the enrichment of allele-specific epigenetic patterns in promoters and regulatory elements (Supplementary Table 8). (c) Read-level methylation and accessibility plots of ZNF597, an imprinted gene with allele-specific epigenetic patterns. (d) Pair-wise comparison of CpG methylation in 1kb regions around heterozygous SV breakpoints between alleles with the SV and alleles without the SV, with significance testing by one-direction Wilcoxon rank-sum tests (Del : 1.3e-6, Ins : 2.0e-5).
