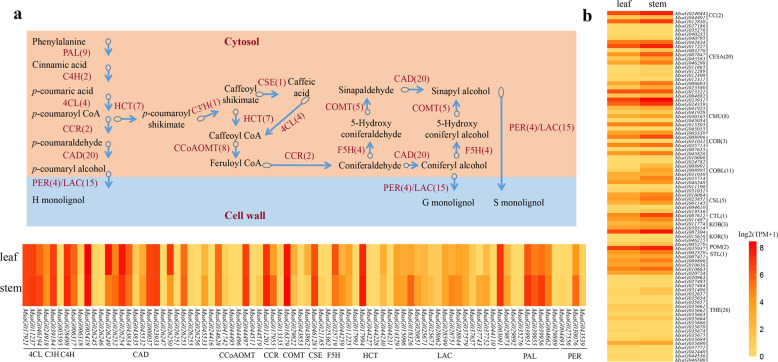
Fig. 6. Expression of lignin and cellulose biosynthesis genes in leaf and stem tissues.
a Simplified diagram of lignin biosynthesis pathways in plants modified from Vanholme et al.78 and a heat map showing the expression of lignin biosynthesis-related genes in leaf and stem tissues. Values within brackets indicate the numbers of gene copies corresponding to the catalytic genes in the pathways. The enzymes involved in the lignin biosynthesis pathway are phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), cinnamoyl-CoA reductase (CCR), cinnamyl alcohol dehydrogenase (CAD), hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase (HCT), p-coumaroyl shikimate/quinate 3’-hydroxylase (C3’H), caffeoyl-CoA O-methyltransferase (CCoAOMT), ferulate 5-hydroxylase (F5H), caffeic acid O-methyltransferase (COMT), peroxidase (PER), laccase (LAC) and caffeoyl shikimate esterase (CSE). b Heat map showing the expression of cellulose-related genes in leaf and stem tissues. The enzymes involved in cellulose biosynthesis pathways are cellulose synthase (CESA), korrigan (KOR), chitinase-like (CTL), cobra (COB), cobra-like (COBL), kobito (KOB), stello (STL), cellulose synthase-microtubule uncoupling protein (CMU), companion of cellulose synthase (CC), theseus (THE), cellulose synthase interacting (POM), and tracheary element differentiation-related (TED).