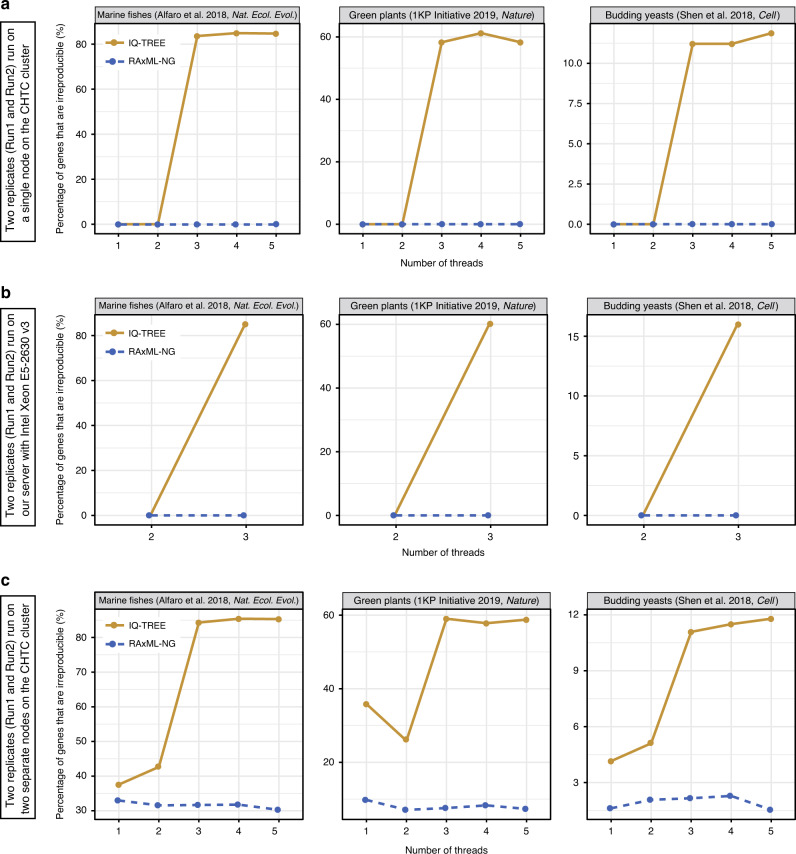
Fig. 5. Variation in computing resources affects the irreproducibility of single-gene trees.
Three large representative studies in animals (marine fishes: 1001 genes and 120 taxa from Alfaro et al.65), plants (green plants: 410 genes and 1178 taxa from 1KP Initiative66), and fungi (budding yeasts: 2408 genes and 343 taxa from Shen et al.16) were used to examine the impact of threading and processor types on the reproducibility of gene phylogenies. Percentages of the 3819 genes whose phylogenies are irreproducible when a we ran two replicates (Run1 and Run2) on a single node (the two replicates were run one right after the other on the same node) on an increasing number of threads on the CHTC cluster using IQ-TREE (in yellow) and RAxML-NG (in blue), respectively. Since running all 3819 genes on a single laboratory server was computationally intractable, we sampled the first 200 genes from each data set. Percentages of these 600 genes when b we ran two replicates on a laboratory server (Intel Xeon E5–2630 v3 @ 2.40 GHz processor with 16 threads) on an increasing number of threads using IQ-TREE (in yellow) and RAxML-NG (in blue), respectively. Percentages of the 3819 genes whose phylogenies are irreproducible when c we ran two replicates (Run1 and Run2) on two separate nodes (i.e., each analysis was run on a single node, but Run1 was executed on a different node than Run2) on an increasing number of threads on the CHTC cluster using IQ-TREE (in yellow) and RAxML-NG (in blue), respectively. The irreproducibility of each gene was determined by comparing the topologies of single-gene trees generated by two replicates (Run1 and Run2). These results suggest that multithreading, coupled with the use of different nodes, is a major contributing factor to irreproducibility in IQ-TREE and that the use of different nodes, but not multithreading, is a major contributing factor to irreproducibility in RAxML-NG. Command lines and job submission files are given in Supplementary Note 1. All gene trees, log files, and statistics of the results, are available on the figshare repository: 10.6084/m9.figshare.11917770.