Fig. 7. An overview of the reproducibility of phylogenetic inference.
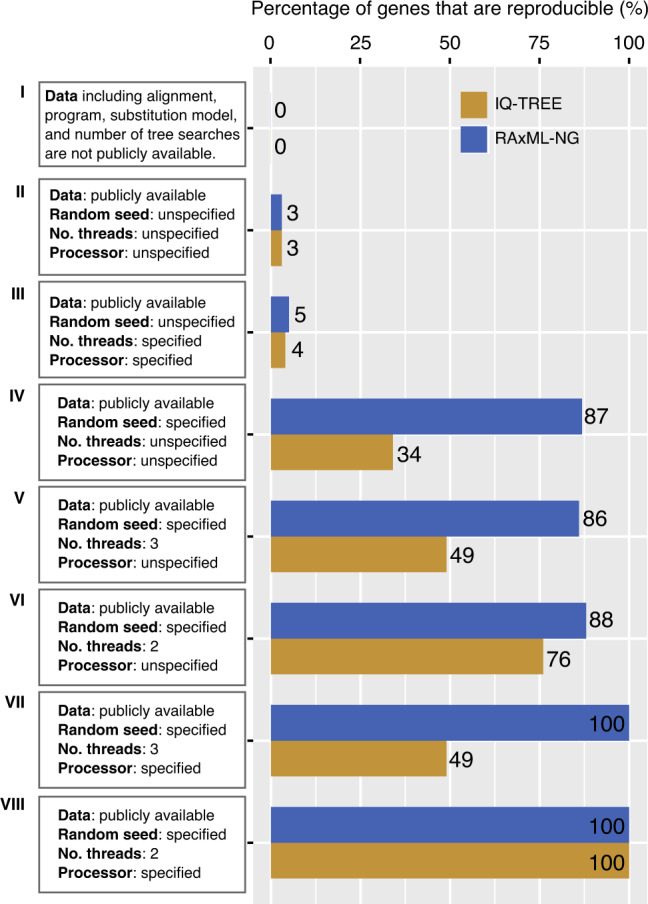
The reproducibility of phylogenetic inference for eight specific scenarios: (I) The data and standard parameter settings typically reported in publications, including sequence alignment, program, substitution model, and a number of tree searches are not publicly available. (II) Sequence alignment, program, substitution model, and a number of tree searches are publicly available, but the number of threads, random starting seed number, and processor are not. (III) Sequence alignment, program, substitution model, number of tree searches, number of threads, and processor are publicly available, but random starting seed number is not. (IV) Sequence alignment, program, substitution model, number of tree searches, and random starting seed number are publicly available, but the number of threads and processors are not. (V) Sequence alignment, program, substitution model, number of tree searches, number of threads (3), and random starting seed number are publicly available, but the processor is not. (VI) Same scenario as V, but with two threads instead of three threads. (VII) Sequence alignment, program, substitution model, number of tree searches, number of threads (3), random starting seed number, and processor are publicly available. (VIII) Same scenario as VII, but with two threads instead of three threads. Analyses for each scenario utilized the first 200 genes from each of three large representative studies in animals (marine fishes: 1001 genes and 120 taxa from Alfaro et al.65), plants (green plants: 410 genes and 1178 taxa from 1KP Initiative66), and fungi (budding yeasts: 2408 genes and 343 taxa from Shen et al.16). Each gene’s reproducibility of phylogenetic inference was assessed using two replicates (Run1 and Run2) for IQ-TREE (in yellow) and RAxML-NG (in blue), respectively. All analyses were performed on the CHTC cluster.
