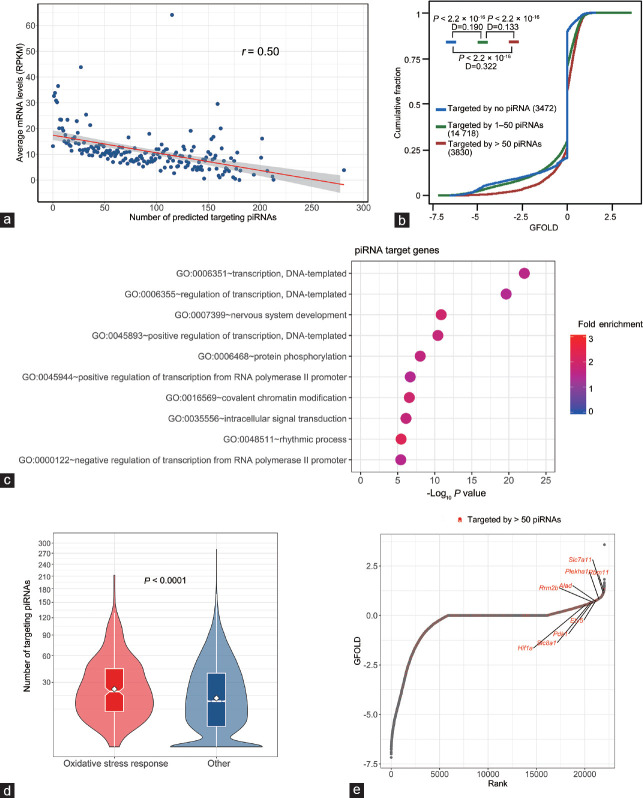
Figure 6.
Expression and function analysis of the potential target genes of epididymal piRNAs. (a) The levels of mRNAs in aged WT caput epididymidis are inversely correlated with the numbers of potential binding piRNAs to their 3'UTRs. The X-axis represents the number of potential binding piRNAs. The Y-axis shows the average levels of mRNAs (RPKM) with the same number of binding piRNAs. (b) Comparison of the cumulative abundance (log10) among the three groups of mRNAs with >50 targeting piRNAs (red line), 1–50 targeting piRNAs (green line), and no apparent targeting piRNAs (blue line). The number of mRNAs in each group is indicated, and P and D values determined by Kolmogorov–Smirnov (KS) test are shown. (c) GO enrichment result for the 3830 genes (Group 1) that are targeted by >50 piRNAs. (d) The number of targeting piRNAs to the set of “oxidative stress response genes” and other genes. The significance of the difference was determined from the Mann–Whitney–Wilcoxon test. (e) Rank-ordered depiction of the GFOLD scores for each gene. The 3830 genes (Group 1) that are targeted by >50 piRNAs are depicted in red. RPKM: reads per kilobase of transcript per million mapped reads; GFOLD: generalized fold change; GO: gene ontology; piRNA: P-element-induced wimpy testis (PIWI)-interacting RNA.