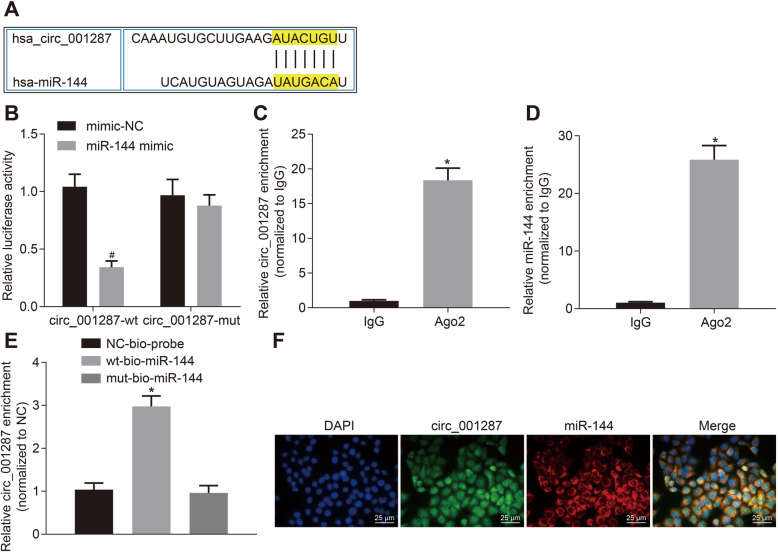
Fig. 4.
miR-144 can specifically bind to circ_001287. a, Binding sites between circ_001287 and miR-144 predicted through a bioinformatics website. b, Binding relationship of miR-144 to circ_001287 measured using dual-luciferase reporter gene assay. c, Binding of circ_001287 and Ago2 measured using RIP assay. d, Binding of miR-144 and Ago2 measured using RIP assay. e, Binding of circ_001287 and miR-144 analyzed using RNA pull-down assay. f, Co-location of circ_001287 and miR-144 in the cytoplasm tested using FISH (× 400). # p < 0.05 vs. the cells treated with mimic-NC; *p < 0.05 vs. IgG and NC-bio-probe. Statistical data were measurement data and described as mean ± standard deviation. Independent sample t-test was used for data comparison between the two groups. Data among groups were compared with one-way ANOVA together with Tukey’s post hoc test. The cell experiment was repeated 3 times. circRNA, circular RNA; miR-144, microRNA-144; Ago2, Argonaut-2; RIP, RNA immunoprecipitation; FISH, Fluorescence in situ hybridization