Figure 2.
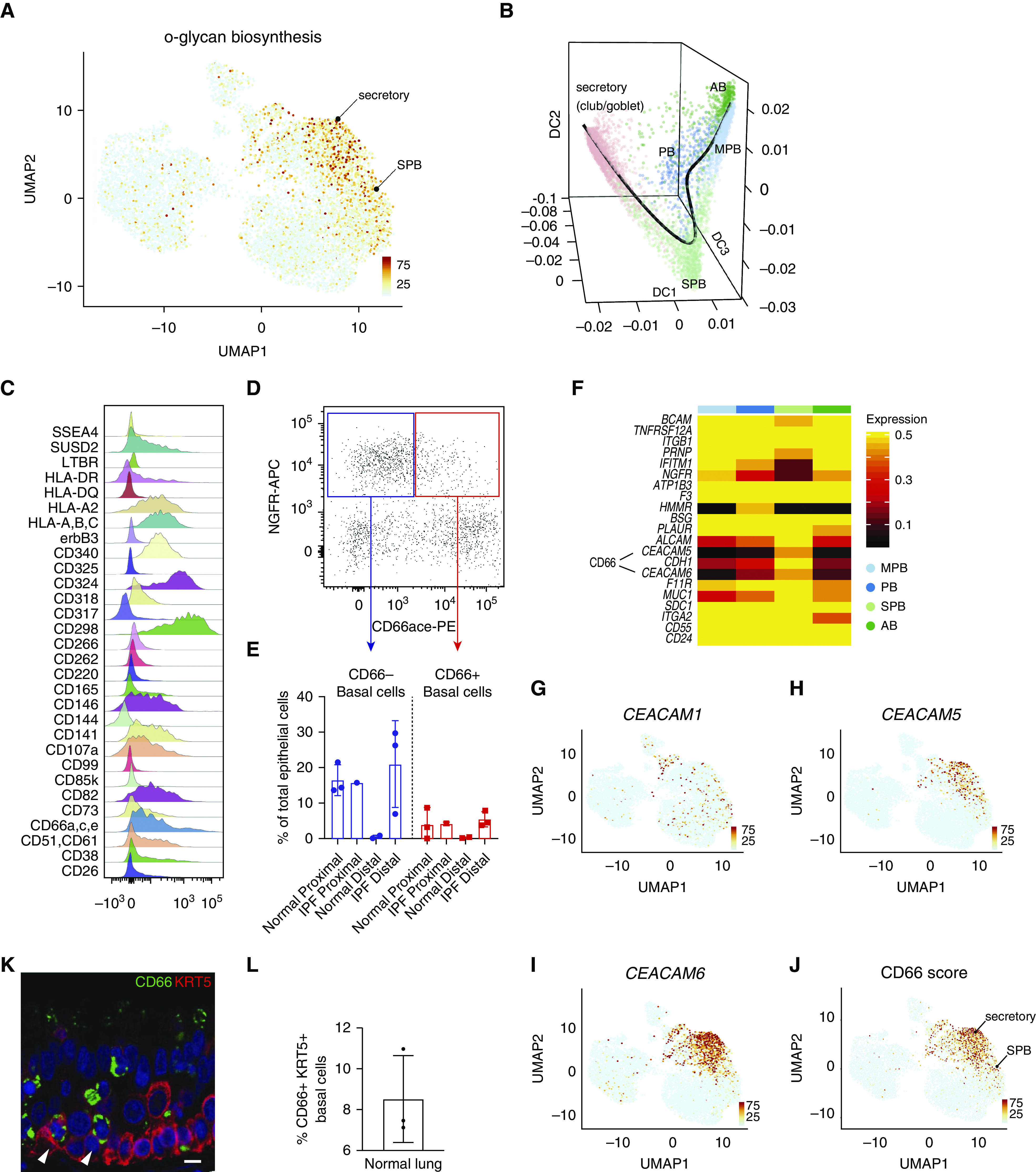
Characterization of secretory primed basal cells (secretory primed basal cluster). (A) Scoring of O-glycan biosynthesis pathway in the normal human airways visualized by Uniform Manifold Approximation and Projection. (B) Lineage reconstruction from basal to secretory cells (black line) was produced with “Slingshot” on the top of a diffusion map generated with “Destiny.” (C) Distribution analysis of CD markers discriminating basal cell subclusters identified by high-content flow cytometry. (D) Discrimination of basal cell subsets by NGFR and CD66 surface reactivity. (E) Percentage of basal subsets over total epithelial cells in proximal and distal compartments of normal and idiopathic pulmonary fibrosis lungs. (F) Differential expression of basal cell–specific CD markers from the single-cell RNA-sequencing data of the four basal cell subclusters. The entire repertoire of human CD molecules available from Human Genome Organization Gene Nomenclature Committee was screened. (G–J) Expression of CEACAM1, CEACAM5, and CEACAM6, and summarized signature score of CD66 in human normal airway epithelium visualized by Uniform Manifold Approximation and Projection. (K) Normal lung cross-section shows immunoreactivity of basal cell subpopulations for CD66 (arrowheads). Scale bar, 15 μm. (L) Quantification of CD66+ basal cells in normal lung, n = 3. AB = activated basal; DC = diffusion component; IPF = idiopathic pulmonary fibrosis; MPB = multipotent basal; NGFR-APC = NGFR allophycocyanin; PB = proliferating basal; PE = phycoerythrin; SPB = secretory primed basal; UMAP = Uniform Manifold Approximation and Projection.
