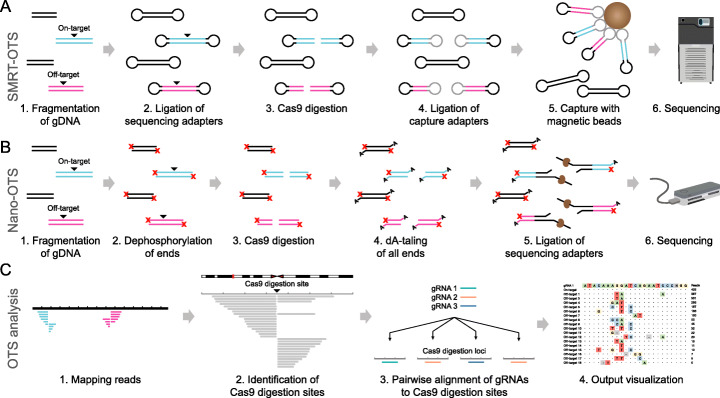
Fig. 1.
The SMRT-OTS and Nano-OTS methods for Cas9 cleavage detection. a SMRT-OTS starts from a DNA sample that has been fragmented by random shearing. The DNA fragments will either contain an on-target site (blue), an off-target (pink), or no gRNA binding site (black) (1). After fragmentation, a SMRTbell library is prepared by ligating sequencing adapters to both ends (2). The SMRTbells containing gRNA binding sites are then cleaved by Cas9 (3), after which capture adapters (gray) are ligated to the cleaved molecules (4). Finally, magnetic beads are used to capture SMRTbells containing the capture adapter (5). This gives an enrichment of SMRTbells cleaved by Cas9 and the enriched library is sequenced on the PacBio Sequel system (6). b Nano-OTS, just like SMRT-OTS, starts from a randomly fragmented DNA sample (1). All ends of the fragmented DNA molecules are dephosphorylated to block adapter ligation (2). The DNA fragments containing gRNA binding sites are cleaved by Cas9 (3) and all 3′ ends are dA-tailed (4). Sequencing adapters are then ligated to Cas9 cleaved and dA-tailed ends (5) and these molecules are sequenced on the ONT MinION system (6). c The same computational approach is used both for SMRT-OTS and Nano-OTS. Reads are aligned to a reference genome, which gives rise to specific patterns at Cas9 cleavage sites with multiple reads starting at the same position (1). The in-house developed software Insider searches for such patterns in the alignment file and reports all detected targets in a bed file (2). The reference sequence is extracted for all the targets and aligned to the gRNA sequences. Only regions with sufficient similarity to a gRNA sequence are kept (3). The results are reported in an output file that contains the Cas9 cleavage position, the sequence alignment of the gRNA to the reference, and the peak height from the OTS sequencing (4)