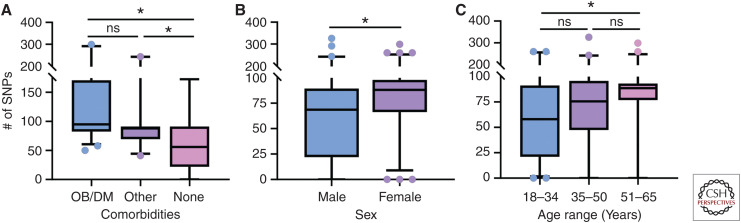
Figure 1.
Increased diversity in viruses derived from high-risk hosts. Using the influenza research database, human surveillance influenza A viral samples with clinical metadata were pooled to look at single-nucleotide polymorphisms (SNPs) in various high-risk groups. The same data set was stratified based on (A) comorbid state of obesity or diabetes mellitus, other comorbidity, or healthy adult with no listed comorbidities, (B) sex, and (C) age range on presentation of symptoms of 18–34, 35–50, or 51–65. Number of SNPs for each gene segment was summed and plotted using box-and-whisker plots ranging from the 5th to 95th percentile with data outside the range represented by data points. Data were analyzed first for normality using the Kolmogorov–Smirnov test, and then for differences in SNPs using a Kruskal–Wallis test (A,C) to compare more than two groups and a Mann–Whitney test (B) to compare two groups. (*) P < 0.05. Database accessed on 30-July-2019.